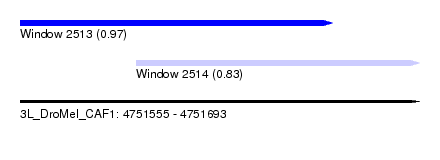
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,751,555 – 4,751,693 |
| Length | 138 |
| Max. P | 0.970233 |

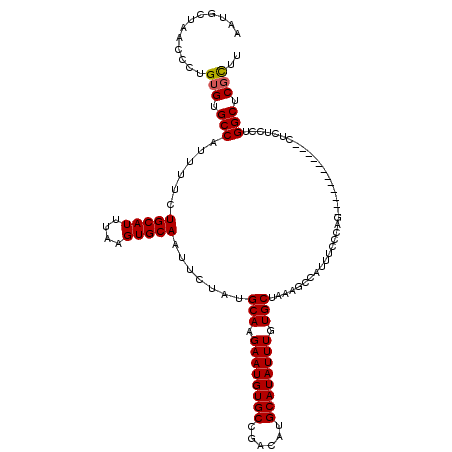
| Location | 4,751,555 – 4,751,663 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.77 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4751555 108 + 23771897 AAUGCUAACCCUGUGUGCCAUUUUCUGCAUUUAAGUGCAAUUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUU ............(((.((((.....(((((....))))).......(((.((((((((......)))))))).)))................------------......)))).))).. ( -25.80) >DroSec_CAF1 11652 108 + 1 AAUGCUAACCCUGUGUGCCAUUUUCUGCAUUUAAGUGCAAUUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUU ............(((.((((.....(((((....))))).......(((.((((((((......)))))))).)))................------------......)))).))).. ( -25.80) >DroSim_CAF1 12297 108 + 1 AAUGCUAACCCUGUGUGCCAUUUUCUGCAUUUAAGUGCAAUUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUU ............(((.((((.....(((((....))))).......(((.((((((((......)))))))).)))................------------......)))).))).. ( -25.80) >DroEre_CAF1 4213 108 + 1 UAUGCUAACCCUGUGUGCCAUUUUCUGCAUUUAAGUGCAAUUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAA------------CUCUCCUGGCUCGCUU ............(((.((((.....(((((....))))).......(((.((((((((......)))))))).)))................------------......)))).))).. ( -25.80) >DroYak_CAF1 10644 108 + 1 UAUGCUAACCCUGUGUGCCAUUUUCUGCAUUUAAGUGCAAUUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUU ............(((.((((.....(((((....))))).......(((.((((((((......)))))))).)))................------------......)))).))).. ( -25.80) >DroAna_CAF1 4486 117 + 1 CUCCC---UCCAGUGUGCCAUUUUCUGCAUUUAAGUGCAAUUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCUCCCCCCAAGGUACUCCCGCCCGGCUCCUUU .....---....(.(((((......(((((....))))).......(((.((((((((......)))))))).)))......................))))).)((....))....... ( -24.00) >consensus AAUGCUAACCCUGUGUGCCAUUUUCUGCAUUUAAGUGCAAUUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG____________CUCUCCUGGCUCGCUU ............(((.(((......(((((....))))).......(((.((((((((......)))))))).)))...................................))).))).. (-24.35 = -24.38 + 0.03)

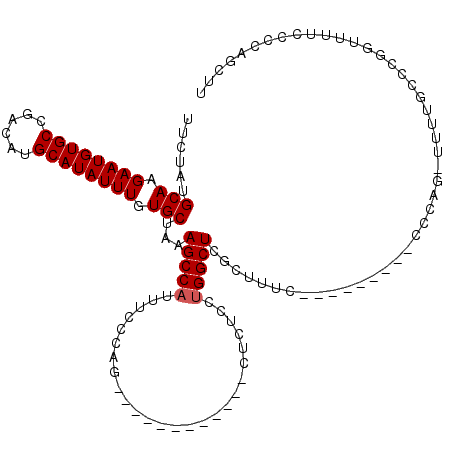
| Location | 4,751,595 – 4,751,693 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4751595 98 + 23771897 UUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUUUC---------CCCAG-UUUUGCCCGGUUUUCUCCAGCUU ......(((.((((((((......)))))))).)))..............((------------((.....(((..(((...---------...))-)...))).((......)))))). ( -20.30) >DroSec_CAF1 11692 78 + 1 UUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUUUC---------CCCAG---------------------CUU ......(((.((((((((......)))))))).)))...(((((........------------......))))).(((...---------...))---------------------).. ( -17.54) >DroSim_CAF1 12337 99 + 1 UUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUUUC---------CCCAGCUUUUGCCCGGUUUUCCCCAGCUU ......(((.((((((((......)))))))).))).((((((.....((((------------.....))))...(((...---------...)))........))))))......... ( -22.00) >DroEre_CAF1 4253 107 + 1 UUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAA------------CUCUCCUGGCUCGCUUUCCCAGUUUUCCCCAG-UUUUCCCCGGUUUUCUCCAGCUU ......(((.((((((((......)))))))).)))..((((..........------------.....((((...(((.....))).....))))-........((......)).)))) ( -19.10) >DroYak_CAF1 10684 99 + 1 UUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG------------CUCUCCUGGCUCGCUUUC---------CCCAGCUUUUGCCCGAUUUUCCCCAGCUU ......(((.((((((((......)))))))).)))..((((......((((------------.....))))...(((...---------...)))...................)))) ( -19.20) >DroAna_CAF1 4523 99 + 1 UUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCUCCCCCCAAGGUACUCCCGCCCGGCUCCUUUGC---------CACCC------------CUUCGGGAGCCA ......(((.((((((((......)))))))).)))............(((((.....(((......))).(((......))---------)....------------....)))))... ( -26.90) >consensus UUCUAUGCAAGAAUGUGCCGACAUGCAUAUUUGUGCUAAAGCCAUUUCCCAG____________CUCUCCUGGCUCGCUUUC_________CCCAG_UUUUGCCCGGUUUUCCCCAGCUU ......(((.((((((((......)))))))).)))...(((((..........................)))))............................................. (-14.60 = -14.77 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:13 2006