
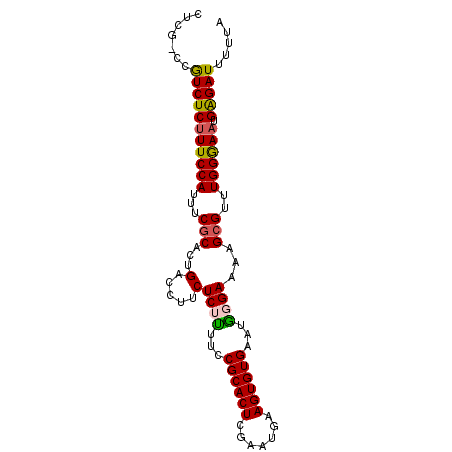
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,750,918 – 4,751,009 |
| Length | 91 |
| Max. P | 0.924672 |

| Location | 4,750,918 – 4,751,009 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -20.16 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4750918 91 + 23771897 AUUG-CCGUCUCAUUCCAUUUCGCACUGACCUUCUCUUUUCCGCACUCGAAUGAAGUGUGAAUGGGAAAAGCGUUUGGGAAUGAGAUUUUUA ....-..((((((((((....(((.......(((((..(((.(((((.......)))))))).)))))..)))....))))))))))..... ( -26.70) >DroSec_CAF1 11036 91 + 1 CUCG-CCGUCUCUUUCCAUUUCUCACUGACCUUCUCUCUUCCGCACUCGAAUGAAGUGUGAAUGGGAAAAGCGUUUGGGAAUGAGAUUUUUA ....-..(((((((((((.........((((((.((((...((((((.......))))))...)))).))).))))))))).)))))..... ( -23.40) >DroSim_CAF1 11672 91 + 1 GUCG-CCGUCUCUUUCCAUUUCGCACUGACCUUCUCUUUUCCGCACUCGAAUGAAGUGUGAAUGGGAAAAGCGUUUGGGAAUGAGAUUUUUA ....-..(((((((((((...(((.......(((((..(((.(((((.......)))))))).)))))..)))..)))))).)))))..... ( -23.20) >DroEre_CAF1 3572 92 + 1 CACUUUCGUCUCUUUCCACUUCGCACUGACCUUCUCUUUUCCGCACUCUAAUGAAGUGUGAAUAUGAAAAGCGCUUGGGAAUGGGAUUUUUA .......(((((((((((...(((...((.....))((((((((((((....).)))))).....))))))))..)))))).)))))..... ( -19.80) >DroYak_CAF1 10001 91 + 1 CUCG-CCAUCUCUUUCCAUUUCGCACUGACCCUCUCUUUUGCGCACUUUAAUGAAGUGUGAAUGUGAAAAGCGUUUGGAAAUGAGAUUUUUA ....-..(((((((((((...(((...(.....)((.....((((((((...)))))))).....))...)))..)))))).)))))..... ( -23.40) >consensus CUCG_CCGUCUCUUUCCAUUUCGCACUGACCUUCUCUUUUCCGCACUCGAAUGAAGUGUGAAUGGGAAAAGCGUUUGGGAAUGAGAUUUUUA .......(((((((((((...(((...(.....)((((...((((((.......))))))...))))...)))..)))))).)))))..... (-20.16 = -20.16 + 0.00)


| Location | 4,750,918 – 4,751,009 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.04 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4750918 91 - 23771897 UAAAAAUCUCAUUCCCAAACGCUUUUCCCAUUCACACUUCAUUCGAGUGCGGAAAAGAGAAGGUCAGUGCGAAAUGGAAUGAGACGG-CAAU ......(((((((((....((((((((((((((...........))))).))))))((.....))...)))....)))))))))...-.... ( -25.70) >DroSec_CAF1 11036 91 - 1 UAAAAAUCUCAUUCCCAAACGCUUUUCCCAUUCACACUUCAUUCGAGUGCGGAAGAGAGAAGGUCAGUGAGAAAUGGAAAGAGACGG-CGAG ......(((((((.((.....((((((((((((...........))))).)))))))....))..)))))))...............-.... ( -21.90) >DroSim_CAF1 11672 91 - 1 UAAAAAUCUCAUUCCCAAACGCUUUUCCCAUUCACACUUCAUUCGAGUGCGGAAAAGAGAAGGUCAGUGCGAAAUGGAAAGAGACGG-CGAC ......((((.((((....((((((((((((((...........))))).))))))((.....))...)))....)))).))))...-.... ( -21.40) >DroEre_CAF1 3572 92 - 1 UAAAAAUCCCAUUCCCAAGCGCUUUUCAUAUUCACACUUCAUUAGAGUGCGGAAAAGAGAAGGUCAGUGCGAAGUGGAAAGAGACGAAAGUG ......(((..(((.((.((.((((((...(((.(((((.....))))).)))...))))))))...)).)))..))).....((....)). ( -20.90) >DroYak_CAF1 10001 91 - 1 UAAAAAUCUCAUUUCCAAACGCUUUUCACAUUCACACUUCAUUAAAGUGCGCAAAAGAGAGGGUCAGUGCGAAAUGGAAAGAGAUGG-CGAG .....(((((.((((((.................(((((.....)))))((((...((.....))..))))...)))))))))))..-.... ( -19.50) >consensus UAAAAAUCUCAUUCCCAAACGCUUUUCCCAUUCACACUUCAUUCGAGUGCGGAAAAGAGAAGGUCAGUGCGAAAUGGAAAGAGACGG_CGAG ......((((.((((((.((.((((((...(((.(((((.....))))).)))...))))))))...))......)))).))))........ (-17.28 = -17.04 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:06 2006