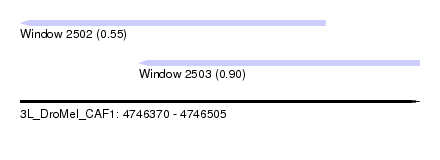
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,746,370 – 4,746,505 |
| Length | 135 |
| Max. P | 0.897812 |

| Location | 4,746,370 – 4,746,473 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -43.34 |
| Consensus MFE | -35.46 |
| Energy contribution | -34.54 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4746370 103 - 23771897 -----GCUUCCUU------------GGAGCUUGGCUCCGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCCUUCUGCGGAUCCAGAAUGGAGGGACGUGAUUUUUUCGUCG -----...(((((------------(((((...)))))))))).(((((((((((.(.((.(((....((....((((((......)))))).))))))).).)))))))))))...... ( -42.90) >DroPse_CAF1 19714 120 - 1 UGGGGGUGUCCUUGCUGGUGACACUGGCACUCGGCUCCGGCGAUAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCCUUCUGUGGAUCCAGAAUGGAGGGACGUGAUUUCUUUGUGG .(..(.(((((((.((((((..((..((((.(((((((.......))))))).))))))..)))))).((((..((((((......))))))...)))))))))))...)..)....... ( -48.30) >DroSec_CAF1 6605 103 - 1 -----GCUUCCUU------------GGAGCUUGGCUCUGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCCUUCUGCGGAUCCAGAAUGGAGGGACGUGAUUUUUUCGUCG -----...((((.------------.((((...))))..)))).(((((((((((.(.((.(((....((....((((((......)))))).))))))).).)))))))))))...... ( -41.20) >DroSim_CAF1 7167 103 - 1 -----GCUUCCUU------------GGAGCUUGGCUCUGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCCUUCUGCGGAUCCAGAAUGGAGGGACGUGAUUUUUUCGCCG -----((.((((.------------.((((...))))..)))).(((((((((((.(.((.(((....((....((((((......)))))).))))))).).)))))))))))..)).. ( -42.30) >DroYak_CAF1 5421 103 - 1 -----GCGUCCUU------------GGAGCUUGGCUCCGGCGACAGGAGUCGCGUGCGUCGCAUCAGGUCAUAGGGAUCCUUCUGCGGAUCCAGAAUGGAGGGACGUGAUUUCUUUGUCG -----........------------(((((...)))))(((((.(((((((((((.(.((.(((....((....((((((......)))))).))))))).).)))))))))))))))). ( -42.00) >consensus _____GCUUCCUU____________GGAGCUUGGCUCCGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCCUUCUGCGGAUCCAGAAUGGAGGGACGUGAUUUUUUCGUCG .....(((((...............))))).((((((((....).)))))))...(((((.(..((..((....((((((......)))))).)).))..).)))))............. (-35.46 = -34.54 + -0.92)


| Location | 4,746,410 – 4,746,505 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -44.74 |
| Consensus MFE | -34.47 |
| Energy contribution | -34.75 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4746410 95 - 23771897 ----CCGUUUCACUGCGACUACGUGACUUGCUGUCC---------GCUUCCUU------------GGAGCUUGGCUCCGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCC ----.........((((((((((((((((.((((((---------.(......------------(((((...)))))).)))))))))))))))).))))))...((((.....)))). ( -43.40) >DroPse_CAF1 19754 117 - 1 GUGGCCGUUUC---GCGGCUACGUGACUUUCCCUCCUUGCUGGGGGUGUCCUUGCUGGUGACACUGGCACUCGGCUCCGGCGAUAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCC (((((((....---.)))))))(((((...(((........)))((((((((....)).)))))).((((.(((((((.......))))))).)))))))))....((((.....)))). ( -54.70) >DroSec_CAF1 6645 95 - 1 ----CCGUUUCACUGCGACUACGUGACUUGCUGUCC---------GCUUCCUU------------GGAGCUUGGCUCUGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCC ----.........((((((((((((((((.((((((---------.(......------------(((((...)))))).)))))))))))))))).))))))...((((.....)))). ( -41.70) >DroSim_CAF1 7207 95 - 1 ----CCGUUUCACUGCGACUACGUGACUUGCUGUCC---------GCUUCCUU------------GGAGCUUGGCUCUGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCC ----.........((((((((((((((((.((((((---------.(......------------(((((...)))))).)))))))))))))))).))))))...((((.....)))). ( -41.70) >DroYak_CAF1 5461 95 - 1 ----CCGUUUCGCUGCGACUACGUGACUUGCUGUCC---------GCGUCCUU------------GGAGCUUGGCUCCGGCGACAGGAGUCGCGUGCGUCGCAUCAGGUCAUAGGGAUCC ----.......(.((((((((((((((((.(((((.---------((.....(------------(((((...))))))))))))))))))))))).)))))).).((((.....)))). ( -42.20) >consensus ____CCGUUUCACUGCGACUACGUGACUUGCUGUCC_________GCUUCCUU____________GGAGCUUGGCUCCGGGGACAGGAGUCGCGUGCGUCGCAUCAGGUCGUAGGGAUCC ..............(((((((((((((((.((((((.........(((((...............)))))..........)))))))))))))))).)))))....((((.....)))). (-34.47 = -34.75 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:02 2006