| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,746,064 – 4,746,163 |
| Length | 99 |
| Max. P | 0.950757 |

| Location | 4,746,064 – 4,746,163 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -44.16 |
| Consensus MFE | -28.24 |
| Energy contribution | -29.72 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4746064 99 + 23771897 ACGGAACUGUUCGUGGUAUCCCGGCCCGCCCAUCCUACGGAUAUCA---------------UCCACAUGCUGCUGCCAUUUUCGCAGCA-ACGGAGGGAGUGCCGGGAACCA-----CAG ............(((((.(((((((.(.(((.(((...((((...)---------------)))....(.((((((.......))))))-.))))))).).)))))))))))-----).. ( -45.40) >DroPse_CAF1 19363 120 + 1 AUGGCACUGUGCGGGGCAUACCGGCGCGGCCCUCCUACGGCUACCAUCACCAUCACCAGCAUCCCCAUGCGGCGGCCAUCUUUGCCACGGACGGGGGCGGCGACUCGCCCCUGAGGGGGG ......((((((.((.....)).))))))(((((((...................((.((((....))))(((((......)))))..)).(((((((((....)))))))))))))))) ( -52.60) >DroSec_CAF1 6299 99 + 1 ACGGCACUGUUCGUGGUAUCCCGGCCCGCCCAUCCUACGGAUAUCA---------------UCCACAUGCUGCUGCCAUUUUCGCAGCG-ACAGAGGGAGUGCCGGGAACCA-----CAG ............(((((.(((((((.(.(((.......((((...)---------------)))...((.((((((.......))))))-.))..))).).)))))))))))-----).. ( -42.10) >DroSim_CAF1 6861 99 + 1 ACGGCACUGUUCGUGGUAUCCCGGCCCGCCCAUCCUACGGGUAUCA---------------UCCACAUGCUGCUGCCAUUUUCGCAGCG-ACUGAGGGAGUGCCGGGAACCA-----CAG ............(((((.(((((((.(.(((..((....)).....---------------....((.(.((((((.......))))))-.))).))).).)))))))))))-----).. ( -41.50) >DroYak_CAF1 5112 99 + 1 AUGGAACGGUGCGUGGCAUCCCGGCUCGCCCAUCCUACGGAUAUCA---------------UCCACAUGCUGCUGCCAUUUUCGCGGCA-ACGGAGGGAGUGCCGGGCACAA-----CAG ........((((.(((((((((((....))..(((...((((...)---------------)))....(.((((((.......))))))-.)))))))).))))).))))..-----... ( -39.20) >consensus ACGGCACUGUUCGUGGUAUCCCGGCCCGCCCAUCCUACGGAUAUCA_______________UCCACAUGCUGCUGCCAUUUUCGCAGCG_ACGGAGGGAGUGCCGGGAACCA_____CAG ......(((....((((.(((((((.(.(((((((...))))...................(((...((((((..........))))))...)))))).).))))))))))).....))) (-28.24 = -29.72 + 1.48)

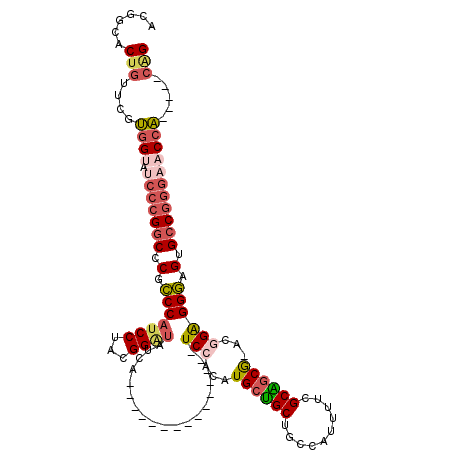

| Location | 4,746,064 – 4,746,163 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -45.74 |
| Consensus MFE | -26.95 |
| Energy contribution | -28.19 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4746064 99 - 23771897 CUG-----UGGUUCCCGGCACUCCCUCCGU-UGCUGCGAAAAUGGCAGCAGCAUGUGGA---------------UGAUAUCCGUAGGAUGGGCGGGCCGGGAUACCACGAACAGUUCCGU .((-----(((((((((((.(.((((((((-((((((.......))))))))....(((---------------(...))))...))).))).).))))))).))))))........... ( -50.50) >DroPse_CAF1 19363 120 - 1 CCCCCCUCAGGGGCGAGUCGCCGCCCCCGUCCGUGGCAAAGAUGGCCGCCGCAUGGGGAUGCUGGUGAUGGUGAUGGUAGCCGUAGGAGGGCCGCGCCGGUAUGCCCCGCACAGUGCCAU .........((((((.(((((((((((((((.(((((.......))))).).))))))..)).))))))((((.((((..((......))))))))))....))))))((.....))... ( -52.60) >DroSec_CAF1 6299 99 - 1 CUG-----UGGUUCCCGGCACUCCCUCUGU-CGCUGCGAAAAUGGCAGCAGCAUGUGGA---------------UGAUAUCCGUAGGAUGGGCGGGCCGGGAUACCACGAACAGUGCCGU .((-----(((((((((((.(.((((((((-.(((((.......))))).))....(((---------------(...))))...))).))).).))))))).))))))........... ( -43.80) >DroSim_CAF1 6861 99 - 1 CUG-----UGGUUCCCGGCACUCCCUCAGU-CGCUGCGAAAAUGGCAGCAGCAUGUGGA---------------UGAUACCCGUAGGAUGGGCGGGCCGGGAUACCACGAACAGUGCCGU .((-----(((((((((((.(.(((((.((-.(((((.......))))).))(((.((.---------------.....)))))..)).))).).))))))).))))))........... ( -42.60) >DroYak_CAF1 5112 99 - 1 CUG-----UUGUGCCCGGCACUCCCUCCGU-UGCCGCGAAAAUGGCAGCAGCAUGUGGA---------------UGAUAUCCGUAGGAUGGGCGAGCCGGGAUGCCACGCACCGUUCCAU ..(-----(.(((((((((.(.((((((((-(((((......))))))).......(((---------------(...))))...))).))).).)))))).))).))............ ( -39.20) >consensus CUG_____UGGUUCCCGGCACUCCCUCCGU_CGCUGCGAAAAUGGCAGCAGCAUGUGGA_______________UGAUAUCCGUAGGAUGGGCGGGCCGGGAUACCACGAACAGUGCCGU .........((((((((((.(.((((((....(((((..........)))))....((......................))...))).))).).))))))).))).............. (-26.95 = -28.19 + 1.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:00 2006