| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,700,098 – 4,700,316 |
| Length | 218 |
| Max. P | 0.698053 |

| Location | 4,700,098 – 4,700,206 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4700098 108 + 23771897 AAACUCAUAAUUAUUUUU--GAAAUUGAACAUGGCAUUUUGAGUUGUCAUUAUCGACUGCAAUUCAGUGCGGUAUGAAUUCAGUAAAGUGAGAACCGAAAAGCAAUAACU ..................--.............((.(((((.(((.(((((....((((.((((((........))))))))))..))))).)))))))).))....... ( -22.60) >DroEre_CAF1 76263 109 + 1 CAAUA-AAAGUUGAAUUUCCGAAAUUGAAAAAGGCGUCUUGAGUUGUCAUUACCGACUGCAAUUCAAUGCGAUAUGAAUUGCAUAUAGUGAGAACCGAGAAGCCAUAACU .....-..(((((..((((.......))))..(((.(((((.(((.((((((.....(((((((((........)))))))))..)))))).)))))))).))).))))) ( -31.80) >DroYak_CAF1 79395 107 + 1 AAAUUUAUACUUAUAUUU--GAAAUUGAAAAAGGCAU-UUGAGCUGUCAUUACCAACUGCAAUUCAAUGCGAUAUAAGCUUAAUAAAAUGAGAACCGAGAAGCCAUAACU ....(((..((((((((.--...((((((....(((.-(((............))).)))..))))))..))))))))..)))........................... ( -13.90) >consensus AAAUU_AUAAUUAUAUUU__GAAAUUGAAAAAGGCAU_UUGAGUUGUCAUUACCGACUGCAAUUCAAUGCGAUAUGAAUUCAAUAAAGUGAGAACCGAGAAGCCAUAACU ................................(((.(((((.(((.(((((......((((......)))).(((.......))).))))).)))))))).)))...... (-14.53 = -14.87 + 0.34)

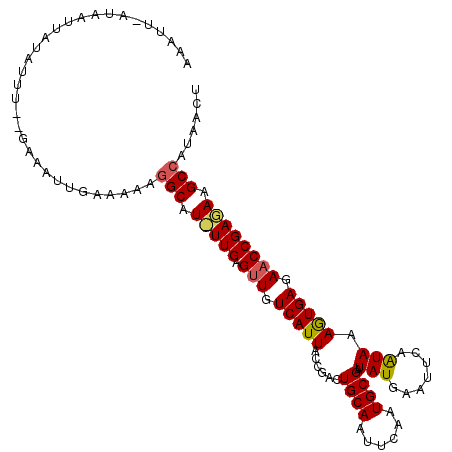
| Location | 4,700,126 – 4,700,245 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.87 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -21.12 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.698053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4700126 119 + 23771897 AUGGCAUUUUGAGUUGUCAUUAUCGACUGCAAUUCAGUGCGGUAUGAAUUCAGUAAAGUGAGAACCGAAAAGCAAUAACUUA-AAACCGCUUAUGGUGUUAUUAAAAAUAGCUUGCGGUU ...((.(((((.(((.(((((....((((.((((((........))))))))))..))))).)))))))).)).........-.((((((....(.((((......)))).)..)))))) ( -32.40) >DroSec_CAF1 73953 120 + 1 CAGCCAUUUUGAGUUUUCAUUAUCGACAGCAUUUCAGUGCGAUAUGAAUUCAAUAAAGUGAGAACCGAGAAGCCUUAACAUAAAAACCGCUUAUGGUGUUAUUAAAAAUAGCUUGCGGUU ..((..(((((.(((((((((.......((((....))))((.......)).....)))))))))))))).))...........((((((....(.((((......)))).)..)))))) ( -26.20) >DroEre_CAF1 76292 119 + 1 AAGGCGUCUUGAGUUGUCAUUACCGACUGCAAUUCAAUGCGAUAUGAAUUGCAUAUAGUGAGAACCGAGAAGCCAUAACUUA-AAACCACUUAUGGUGUUAUUAAAAAUAGCUUGCGGUU ..(((.(((((.(((.((((((.....(((((((((........)))))))))..)))))).)))))))).)))........-.((((.(....(.((((......)))).)..).)))) ( -34.50) >DroYak_CAF1 79423 118 + 1 AAGGCAU-UUGAGCUGUCAUUACCAACUGCAAUUCAAUGCGAUAUAAGCUUAAUAAAAUGAGAACCGAGAAGCCAUAACUUA-AAACCGCUUAUGGUGUUAUUAAAAAUAGCUUGCGGUU ..(((..-(((((((((........))((((......)))).....))))))).....((.....))....)))........-.((((((....(.((((......)))).)..)))))) ( -20.80) >consensus AAGGCAUUUUGAGUUGUCAUUACCGACUGCAAUUCAAUGCGAUAUGAAUUCAAUAAAGUGAGAACCGAGAAGCCAUAACUUA_AAACCGCUUAUGGUGUUAUUAAAAAUAGCUUGCGGUU ..(((.(((((.(((.(((((.......(((......)))..(((.......))).))))).)))))))).)))..........((((((....(.((((......)))).)..)))))) (-21.12 = -21.75 + 0.62)

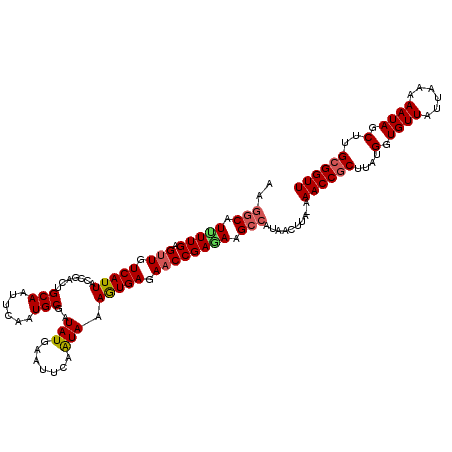
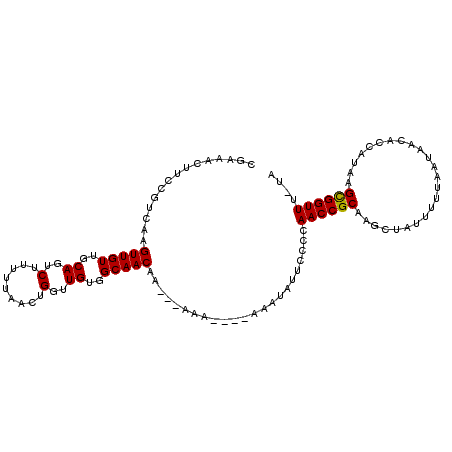
| Location | 4,700,206 – 4,700,316 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.05 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4700206 110 - 23771897 CGAAACUUCCGUCAAGUUGUUGCAGUCUUUUUAACUGGUUGUGGCAACAAAAAAAAAAUAAAAAAUUCCCCAACCGCAAGCUAUUUUUAAUAACACCAUAAGCGGUUU-UA ...............(((((..((..(.........)..))..))))).......................((((((........................)))))).-.. ( -19.46) >DroSec_CAF1 74033 107 - 1 CGAAACUUCCGUCAAGUUGUUGCAGUCUUUUUAACUGGUUGUGGCAACAACAAAAA----AAAAAUUCCCCAACCGCAAGCUAUUUUUAAUAACACCAUAAGCGGUUUUUA ...............(((((..((..(.........)..))..)))))........----...........((((((........................)))))).... ( -19.46) >DroSim_CAF1 75358 102 - 1 CGAAACUUCCGUCAAGUUGUUGCAGUCUUUUUAACUGGUUGUGGCAACA----AAA----AAAUAUUCCCCAACCGCAAGCUAUUUUUAAUAACACCAUAAGCGGUUU-UA ...............(((((..((..(.........)..))..))))).----...----...........((((((........................)))))).-.. ( -19.46) >DroEre_CAF1 76372 101 - 1 CGAAACUUCCGUCAAGUUGUUGCAGUCUUUUUAACUGGUUGUGGCAACAA-----A----AAAUAUUCUCCAACCGCAAGCUAUUUUUAAUAACACCAUAAGUGGUUU-UA ...............(((((..((..(.........)..))..)))))..-----.----...........((((((........................)))))).-.. ( -17.56) >DroYak_CAF1 79502 101 - 1 CGAAACUUCCGUCAAGUUGUUGCAGUCUUUUUAACUGGUUGUGGCAACAA-----A----AAAUAUUCCCCAACCGCAAGCUAUUUUUAAUAACACCAUAAGCGGUUU-UA ...............(((((..((..(.........)..))..)))))..-----.----...........((((((........................)))))).-.. ( -19.46) >consensus CGAAACUUCCGUCAAGUUGUUGCAGUCUUUUUAACUGGUUGUGGCAACAA___AAA____AAAUAUUCCCCAACCGCAAGCUAUUUUUAAUAACACCAUAAGCGGUUU_UA ...............(((((..((..(.........)..))..))))).......................((((((........................)))))).... (-19.24 = -19.08 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:41 2006