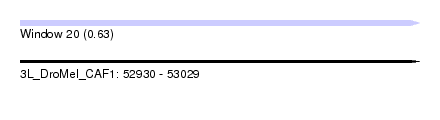
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 52,930 – 53,029 |
| Length | 99 |
| Max. P | 0.633094 |

| Location | 52,930 – 53,029 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -19.83 |
| Energy contribution | -19.47 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.633094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
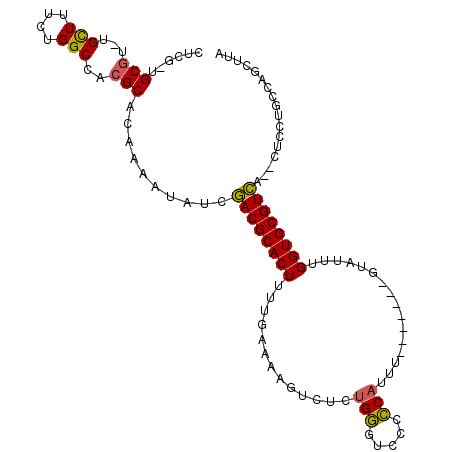
>3L_DroMel_CAF1 52930 99 + 23771897 CUCG-UGCGU-UGCUUUCUGGCUACGCACAUAAUAUCGACGCACUUUUUAAAAGUCUCUGGUUCCACCAUUU-------GUAUUUGGUGCGUCA--CUCCUGCCAGCUUA ...(-(((((-.(((....))).))))))........((((((((......((((...((....))..))))-------......)))))))).--.............. ( -29.00) >DroVir_CAF1 15356 108 + 1 GUUGUCGCUUACGCUUCCUGGCCACGCACAAAAUAUCAACGCACUUUUCAA--GUCCUCGCAUUUCGCAUUUCGUAGUCGUAUUUGGUGCGUUGGCCGCCUUUUAGGUUG ......((....)).....((((((((((.((((((.(..(((((.....)--))....((.....)).....))..).)))))).))))).)))))(((.....))).. ( -26.20) >DroSec_CAF1 2789 99 + 1 CUCGUUGCGU-UGCUUUCUGGCUACGCACAUAAUAUCGACGCACUUUUGAAAAGUCUCUGGGUCU-CCAUUU-------GGAUUUGGUGCGUCA--CUCCUGCCAGCCUA ...((.((((-.(((....))).))))))........((((((((......((((((.(((....-)))...-------)))))))))))))).--.............. ( -27.80) >DroSim_CAF1 2700 100 + 1 CUCGUUGCGU-UGCUUUCUGGCUACGCACAUAAUAUCGACGCACUUUUGAAAAGUCUCUGGGUCCCCCAUUU-------GGAUUUGGUGCGUCA--CUCCUGCCAGCUUA ...((.((((-.(((....))).))))))........((((((((......((((((.((((...))))...-------)))))))))))))).--.............. ( -29.80) >DroEre_CAF1 2787 99 + 1 AACG-UGCGU-UGUUUUCUGGCCACGCACAAAAUAUCGACGCACUUUUGAAAAGUCUCUGGGUCCCCCAUUU-------GUAUUUGGUGCGUCA--UUCCUACCAGCUUG ...(-(((((-.(((....))).))))))........((((((((......(((....((((...)))))))-------......)))))))).--.............. ( -27.80) >DroYak_CAF1 2775 99 + 1 CUCG-UGCGU-UGCUUUCUGGCCACGCACAAAAUAUCGACGCACUUUUGAAAAGUCUCUGGGUCCCCCAUUU-------GUAUUUGGUGCGUCA--CUCCUGCCAGCUUG ...(-(((((-.(((....))).))))))........((((((((......(((....((((...)))))))-------......)))))))).--.............. ( -30.30) >consensus CUCG_UGCGU_UGCUUUCUGGCCACGCACAAAAUAUCGACGCACUUUUGAAAAGUCUCUGGGUCCCCCAUUU_______GUAUUUGGUGCGUCA__CUCCUGCCAGCUUA ......(((...(((....)))..)))..........((((((((.............(((.....)))................))))))))................. (-19.83 = -19.47 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:57 2006