
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,680,294 – 4,680,397 |
| Length | 103 |
| Max. P | 0.937933 |

| Location | 4,680,294 – 4,680,397 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
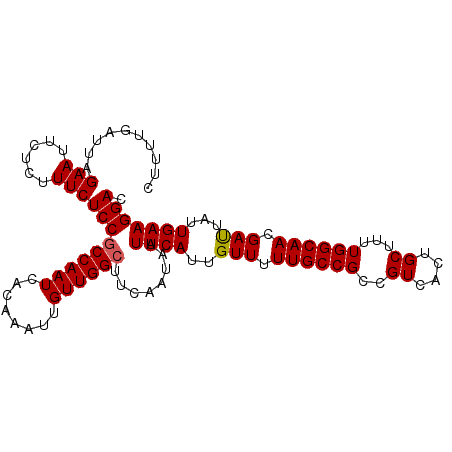
>3L_DroMel_CAF1 4680294 103 + 23771897 CUUUUGAUUAGAAUUCUCUUUCUCCGCCAAUCACAAAUUGUUGGCAACGAUAAUUCAUUGUUUUUGCCGCCGUCACUGCUUUUGGCAACGAUUAUUGAAGGAC ((((..((.((((......))))............((((((((((((.(((((....))))).)))))((((..........)))))))))))))..)))).. ( -22.70) >DroSec_CAF1 52345 103 + 1 CUUUUGAUUAGAAUUCUCUUUCUCCGCCAAUCACAAAUUGUUGGCUUCAAUAAUUCAUUGUUUUUGCCGCCGUCACUGCUUUUGGCAACGAUUAUUGAAGGAC ..........(((......)))(((((((((........))))))((((((((((..(((((...((.(......).))....))))).))))))))))))). ( -21.90) >DroSim_CAF1 55411 103 + 1 CUUUUGAUUAGAAUUCUCUUUCUCCGCCAAUCACAAAUUGUUGGCUUCAAUAAUUCAUUGUUUUUGCCGCCGUCACUGCUUUUGGCAACGACUAUUGAAGGAC ..........(((......)))(((((((((........))))))........((((..(((.((((((..((....))...)))))).)))...))))))). ( -21.70) >DroEre_CAF1 56314 103 + 1 CUUUUGAUUAGAAUUCUCUUUCUCCGCCAAUCACAAAUUGUUGGCUUCAAUAAUUCAUUGUUUUUGCCGCCGUCACUGCUUUUGGCAACGAUUAUUGAAGGAC ..........(((......)))(((((((((........))))))((((((((((..(((((...((.(......).))....))))).))))))))))))). ( -21.90) >DroYak_CAF1 59069 103 + 1 CUUUUGAUUAGAAUUCUCUUUCUCCGCCAAUCACAAAUUGUUGGCUUCAAUAAUUCAUUGUUUUUGCCGCCGUCACUGCUUUUGGCAACGAUUAUUGAAGGAC ..........(((......)))(((((((((........))))))((((((((((..(((((...((.(......).))....))))).))))))))))))). ( -21.90) >DroAna_CAF1 54813 103 + 1 GUUCUGAUUAGAAUUCUCUUUCUCCUCCAAUCACAAUUUGUUGGCUUCAAUAAUUCAUUGUUUUUGCCGCCGUCACUGCUUUUGGCAACGAUUAUUGAAGGAC (((((((..((((......))))..))...(((....((((((((...(((((....)))))...)))((((..........)))))))))....)))))))) ( -18.90) >consensus CUUUUGAUUAGAAUUCUCUUUCUCCGCCAAUCACAAAUUGUUGGCUUCAAUAAUUCAUUGUUUUUGCCGCCGUCACUGCUUUUGGCAACGAUUAUUGAAGGAC ..........(((......)))(((((((((........))))))........((((..(((.((((((..((....))...)))))).)))...))))))). (-19.52 = -19.55 + 0.03)

| Location | 4,680,294 – 4,680,397 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 97.41 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.58 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4680294 103 - 23771897 GUCCUUCAAUAAUCGUUGCCAAAAGCAGUGACGGCGGCAAAAACAAUGAAUUAUCGUUGCCAACAAUUUGUGAUUGGCGGAGAAAGAGAAUUCUAAUCAAAAG ............((.(((((((..(((.((..((((((.................))))))..))...)))..))))))).)).................... ( -20.43) >DroSec_CAF1 52345 103 - 1 GUCCUUCAAUAAUCGUUGCCAAAAGCAGUGACGGCGGCAAAAACAAUGAAUUAUUGAAGCCAACAAUUUGUGAUUGGCGGAGAAAGAGAAUUCUAAUCAAAAG .(((((((((((((((((......((.((....)).)).....))))).)))))))))(((((((.....)).))))))))((...((....))..))..... ( -22.80) >DroSim_CAF1 55411 103 - 1 GUCCUUCAAUAGUCGUUGCCAAAAGCAGUGACGGCGGCAAAAACAAUGAAUUAUUGAAGCCAACAAUUUGUGAUUGGCGGAGAAAGAGAAUUCUAAUCAAAAG .(((((((((((((((..(........)..)))))(.......).......)))))))(((((((.....)).))))))))((...((....))..))..... ( -23.50) >DroEre_CAF1 56314 103 - 1 GUCCUUCAAUAAUCGUUGCCAAAAGCAGUGACGGCGGCAAAAACAAUGAAUUAUUGAAGCCAACAAUUUGUGAUUGGCGGAGAAAGAGAAUUCUAAUCAAAAG .(((((((((((((((((......((.((....)).)).....))))).)))))))))(((((((.....)).))))))))((...((....))..))..... ( -22.80) >DroYak_CAF1 59069 103 - 1 GUCCUUCAAUAAUCGUUGCCAAAAGCAGUGACGGCGGCAAAAACAAUGAAUUAUUGAAGCCAACAAUUUGUGAUUGGCGGAGAAAGAGAAUUCUAAUCAAAAG .(((((((((((((((((......((.((....)).)).....))))).)))))))))(((((((.....)).))))))))((...((....))..))..... ( -22.80) >DroAna_CAF1 54813 103 - 1 GUCCUUCAAUAAUCGUUGCCAAAAGCAGUGACGGCGGCAAAAACAAUGAAUUAUUGAAGCCAACAAAUUGUGAUUGGAGGAGAAAGAGAAUUCUAAUCAGAAC ...(((((((((((((((......((.((....)).)).....))))).))))))))))........((.(((((((((...........))))))))).)). ( -23.50) >consensus GUCCUUCAAUAAUCGUUGCCAAAAGCAGUGACGGCGGCAAAAACAAUGAAUUAUUGAAGCCAACAAUUUGUGAUUGGCGGAGAAAGAGAAUUCUAAUCAAAAG .(((((((((((((((((......((.((....)).)).....))))).)))))))))(((((((.....)).))))))))((...((....))..))..... (-20.06 = -20.58 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:34 2006