
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,647,584 – 4,647,732 |
| Length | 148 |
| Max. P | 0.937710 |

| Location | 4,647,584 – 4,647,692 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -41.16 |
| Consensus MFE | -29.31 |
| Energy contribution | -28.23 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
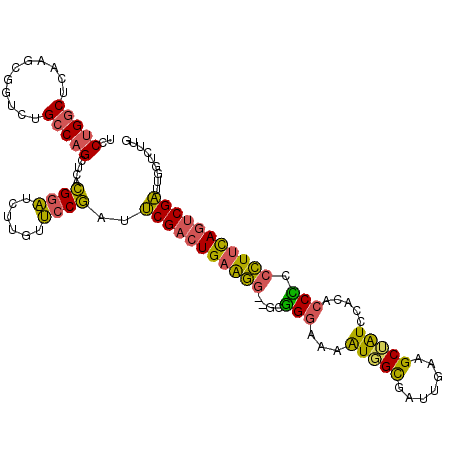
>3L_DroMel_CAF1 4647584 108 + 23771897 UCCUGGCUCAAGCGAUCUGCCAGCUCACGGAUUUUAUUCCGAUUCGACUGAAGG--GCGGGAAAAUGGCGAUUGAAGCUAUCCACACCCCCCUUUAGUCGAUUGGUCUUG ..(((((...........)))))....((((......))))..(((((((((((--(.(((...(((((.......))))).....)))))))))))))))......... ( -42.10) >DroSec_CAF1 20010 108 + 1 UCCUGGCUCAAGCGGUCUGCCAGCUCACGGAUCUUGUUCCGAUUCGACUGAAGG--GCGGGAAAAUGGCGAUUGAAGCUAUCCACACCUCCCUUUAGUCGAUUGGUCUUG ..(((((...........)))))....((((......))))..(((((((((((--(.((....(((((.......))))).....)).))))))))))))......... ( -40.80) >DroSim_CAF1 20585 108 + 1 UCCUGGCUCAAGCGGUCUGCCAGCUCACGGAUCUUGUUCCGAUUCGACUGAAGG--GCGGGAAAAUGGCGAUUGAAGCUAUCCACACCUCCCUUUAGUCGAUUGGUCUUG ..(((((...........)))))....((((......))))..(((((((((((--(.((....(((((.......))))).....)).))))))))))))......... ( -40.80) >DroEre_CAF1 20232 108 + 1 UCCUGGCUCAGGCGGUCUGCCAGUUCGCGGAUCUUGUUCCGAUUCGACUGAAGG--GCGGGAAAAUGGCGGUUGAAGCUAUCCACGCCCCCCUUCAGUCGAUUGGUCUUG .((((...)))).(((((((......))))))).....(((((.((((((((((--(.(((...(((((.......))))).....)))))))))))))))))))..... ( -48.20) >DroYak_CAF1 23487 108 + 1 UCCUGGCUCAAACGGUCUGCCAGCUCACGGAUCUUGUUCCGAUUCGACUGAAGG--GCGGGAAAAUUGCGAUUAAAGCUAUCCACGCCCCCCUUCAGUCGAUUGGUCUUG ..(((((...........)))))....((((......))))..(((((((((((--(.(((......((.......))........)))))))))))))))......... ( -42.04) >DroPer_CAF1 25237 108 + 1 UCCCGACUCAAGCGCUCCGCCUGCUUCAGUUUACUGUGCCUAAGCGA-UGAGUAUCUCUGAAACGUUGU-GUUGAAGCAGUCGACUGCGCCGUGCAGCCGUUGGGUCCUA ....(((((((..(((.(((..((..(((((.(((((..(...((((-((.............))))))-...)..))))).))))).)).))).)))..)))))))... ( -33.02) >consensus UCCUGGCUCAAGCGGUCUGCCAGCUCACGGAUCUUGUUCCGAUUCGACUGAAGG__GCGGGAAAAUGGCGAUUGAAGCUAUCCACACCCCCCUUCAGUCGAUUGGUCUUG ..(((((...........)))))....((((......))))..(((((((((((....(((...(((((.......))))).....))).)))))))))))......... (-29.31 = -28.23 + -1.07)

| Location | 4,647,624 – 4,647,732 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -25.07 |
| Energy contribution | -25.99 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
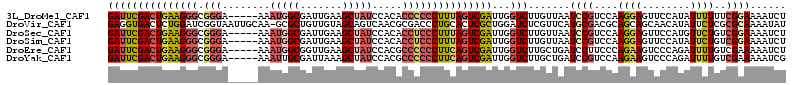
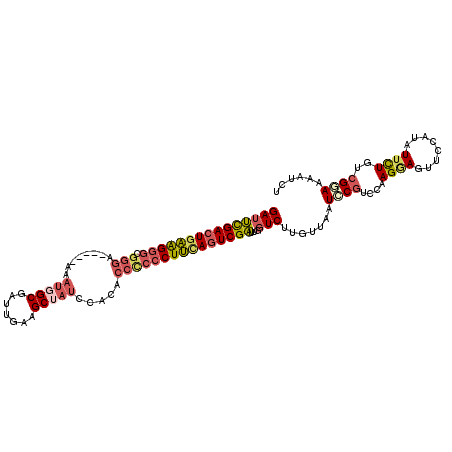
>3L_DroMel_CAF1 4647624 108 + 23771897 GAUUCGACUGAAGGGCGGGA-----AAAUGGCGAUUGAAGCUAUCCACACCCCCCUUUAGUCGAUUGGUCUUGUUAAUCCGUCCAAGGAGUUCCAUAUUUUUUCGGAAAAUCU (((((((((((((((.(((.-----..(((((.......))))).....)))))))))))))))...))).......((((...(((((((.....)))))))))))...... ( -35.70) >DroVir_CAF1 35010 112 + 1 GAGGUGACUCUGGAUCGGUAAUUGCAA-GCGCUGUUGUAGCAGUCAACGCGACCCUGCACUCGCUGGAUCUCGUUCAUGCGACGCAGCAGCAACAUAUUCUCGCGCAAAAUAU (((....)))...........((((..-..((((((((.(((...((((.((.((.((....)).)).)).))))..)))...))))))))..(........).))))..... ( -33.60) >DroSec_CAF1 20050 108 + 1 GAUUCGACUGAAGGGCGGGA-----AAAUGGCGAUUGAAGCUAUCCACACCUCCCUUUAGUCGAUUGGUCUUGUUAAUCCGUCCAAGGAGUUCCAUGUUCUGUCGGAAAAUCU (((((((((((((((.((..-----..(((((.......))))).....)).))))))))))))...))).......((((.(((.((....)).))....).))))...... ( -35.50) >DroSim_CAF1 20625 108 + 1 GAUUCGACUGAAGGGCGGGA-----AAAUGGCGAUUGAAGCUAUCCACACCUCCCUUUAGUCGAUUGGUCUUGUUAAUCCGUCCAAGGAGUUCCAUAUUCUGUCGGAAAAUCU (((((((((((((((.((..-----..(((((.......))))).....)).))))))))))))...))).......((((.....(((((.....)))))..))))...... ( -36.70) >DroEre_CAF1 20272 108 + 1 GAUUCGACUGAAGGGCGGGA-----AAAUGGCGGUUGAAGCUAUCCACGCCCCCCUUCAGUCGAUUGGUCUUGCUGAUCCUUCCCAGAAGUCCCAGAUUUUGUCGAAAAAUCU ...((((((((((((.(((.-----..(((((.......))))).....)))))))))))))))..((.(((.(((........)))))).)).(((((((.....))))))) ( -43.40) >DroYak_CAF1 23527 108 + 1 GAUUCGACUGAAGGGCGGGA-----AAAUUGCGAUUAAAGCUAUCCACGCCCCCCUUCAGUCGAUUGGUCUUGCUGAUCCGUCCAAGAAGUCCCAGAUUUUGUCGAAAAAUCG ...((((((((((((.(((.-----.....((.......))........))))))))))))))).((((((((..((....)))))))....)))((((((.....)))))). ( -37.44) >consensus GAUUCGACUGAAGGGCGGGA_____AAAUGGCGAUUGAAGCUAUCCACACCCCCCUUCAGUCGAUUGGUCUUGUUAAUCCGUCCAAGGAGUUCCAUAUUCUGUCGGAAAAUCU (((((((((((((((.(((........(((((.......))))).....)))))))))))))))...))).......((((....((((........))))..))))...... (-25.07 = -25.99 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:21 2006