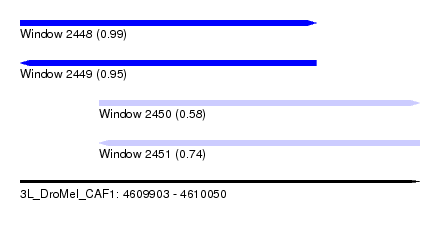
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,609,903 – 4,610,050 |
| Length | 147 |
| Max. P | 0.988147 |

| Location | 4,609,903 – 4,610,012 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4609903 109 + 23771897 -----CCGGUUCGGGAUCAGGUUCCGGUGCC------GGGGCCAAGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUA -----..(.((((((.((.(((((((....)------))))))..((.(((((.(..(((.....)))..).))))).))((((((((((.....))))))))..)))).).))))).). ( -43.80) >DroVir_CAF1 221407 97 + 1 --------------GC---GGCGCCUCGUCG------GCGUCGAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUA --------------(.---((((((.(((((------(((((....))))))..(.....).......)))).)))((.(((((((((((.....))))))))..))).))))))..... ( -44.40) >DroGri_CAF1 207721 111 + 1 UGUGGCAACAACGGGU---GGCGCCUCCGCG------ACUAAGGGGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUA ..((....)).(...(---(((((((((.(.------.....))))))))))).).........(((((....(((((.(((((((((((.....))))))))..))).)))))))))). ( -46.30) >DroWil_CAF1 165548 106 + 1 -----CUGCAGCUGGG---GGCAACUCAUCG------AGCACUAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGUCCGCCCAAGUA -----.(((.(((.((---(....)))....------))).....((((((((.(...............).))))((.((.((((((((.....))))))))...)).))))))..))) ( -32.46) >DroMoj_CAF1 245025 104 + 1 GGUGGCA-------GU---GGUGCCGCCGCU------GCAUCGAAGGGCGCCAAGGAGAACAAUAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUA ((((.((-------((---((.....)))))------)))))...((((((((.(...............).))))((.(((((((((((.....))))))))..))).))))))..... ( -42.46) >DroAna_CAF1 196771 115 + 1 -----CUGGCUCGGGCUCAGGAUCCGGAUCCGGAACUGGCGCCAAGGGCACCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGACCGCCGAAGUA -----...((((.(((((((..((((....)))).)))).)))..)))).......................((((((.((.((((((((.....))))))))...)).))))))..... ( -40.50) >consensus _____CAG___CGGGC___GGCGCCGCAGCG______GCCACGAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUA ...................(((.((.....................)).))).....................(((((.(((((((((((.....))))))))..))).)))))...... (-23.98 = -24.48 + 0.50)


| Location | 4,609,903 – 4,610,012 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -37.91 |
| Consensus MFE | -25.33 |
| Energy contribution | -25.14 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4609903 109 - 23771897 UACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGCCCC------GGCACCGGAACCUGAUCCCGAACCGG----- ......(((((((((..((((((.........)))))))))))))))..(((((............)))))(((...))).((------((..(.(((......))).)..))))----- ( -37.10) >DroVir_CAF1 221407 97 - 1 UAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUCGACGC------CGACGAGGCGCC---GC-------------- ......(((((((((..((((((.........))))))))))))))).....................(((((((.(((....------))).).))))))---..-------------- ( -40.20) >DroGri_CAF1 207721 111 - 1 UAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCCCCUUAGU------CGCGGAGGCGCC---ACCCGUUGUUGCCACA (((((((((((((((..((((((.........)))))))))))))))....))))))..........(((((((.(((.....------.).)).))))))---)............... ( -41.00) >DroWil_CAF1 165548 106 - 1 UACUUGGGCGGACGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUAGUGCU------CGAUGAGUUGCC---CCCAGCUGCAG----- ......(((((.(((..((((((.........))))))))).))))).........((((..(((.(((((((.....)))).------))).))).....---..)))).....----- ( -28.30) >DroMoj_CAF1 245025 104 - 1 UAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUAUUGUUCUCCUUGGCGCCCUUCGAUGC------AGCGGCGGCACC---AC-------UGCCACC ......(((((((((..((((((.........)))))))))))))))..((((..............))))(((((.......------)).)))((((..---..-------))))... ( -35.84) >DroAna_CAF1 196771 115 - 1 UACUUCGGCGGUCGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGUGCCCUUGGCGCCAGUUCCGGAUCCGGAUCCUGAGCCCGAGCCAG----- ......(((((((((..((((((.........)))))))))))))))............((((....(((((((...)))))))((((.((((....)))).))))..))))...----- ( -45.00) >consensus UACUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUCGACCC______CGAGGAGGCGCC___ACCCG___CAG_____ ......(((((((((..((((((.........))))))))))))))).(((((..............)))))................................................ (-25.33 = -25.14 + -0.19)


| Location | 4,609,932 – 4,610,050 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -24.69 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4609932 118 + 23771897 GCCAAGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCGAAGUACAAGCCACCGGAUUUCACCUAGUGUCG--UCUCAUUCCCG .....(((.......((((...(((((.(....(((((.(((((((((((.....))))))))..))).)))))(((((.(........).)))))..).)))))..--))))...))). ( -36.00) >DroVir_CAF1 221424 106 + 1 UCGAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUACAAGCCGCCGGACUUUACCUAGCGCC-------------- ......(((((..(((...............(.(((((.(((((((((((.....))))))))..))).))))))......(((((...)).)))..))).)))))-------------- ( -34.40) >DroGri_CAF1 207752 117 + 1 AAGGGGGGCGCCAAGGAGAACAACAUUUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUAUAAACCGCCCGAUUUUACCUAAUCCCG--GGAAAUUCCG- ..(((((((((((.(..(((.....)))..).))))((.(((((((((((.....))))))))..))).))))))............((((..............))--))....))).- ( -37.64) >DroWil_CAF1 165574 120 + 1 ACUAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGUCCGCCCAAGUACAAGCCGCCAGAUUUUACCUAGCGUCUAACCUAAAUCCUC .....((((((..(((((((((.....))...(((((((...((((((((.....))))))))....((.(......).))..)))))))..)))).))).))))))............. ( -31.90) >DroMoj_CAF1 245049 106 + 1 UCGAAGGGCGCCAAGGAGAACAAUAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUACAAGCCGCCGGAUUUUACCUAGCAUA-------------- .....((((((((.(...............).))))((.(((((((((((.....))))))))..))).))))))...........((..(.......)..))...-------------- ( -30.66) >DroAna_CAF1 196806 118 + 1 GCCAAGGGCACCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAACGACCGCCGAAGUACAAGCCGCCGGACUUCCCCUAGCGGCG--UGUACCUUCCC (((...))).....((((..............((((((.((.((((((((.....))))))))...)).))))))..(((((.(((((..(.......)..))))).--))))).)))). ( -42.30) >consensus ACGAAGGGCGCCAAGGAGAACAACAUAUGACGUGGCGGCCGCAAUUGGUUAUAGAAGCCAAUUAAGCGACCGCCCAAAUACAAGCCGCCGGAUUUUACCUAGCGUCG____UAA_UCC__ ......(((((..(((.(((((.....))....(((((.(((((((((((.....))))))))..))).)))))...................))).))).))))).............. (-24.69 = -25.47 + 0.78)



| Location | 4,609,932 – 4,610,050 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | -30.48 |
| Energy contribution | -30.09 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4609932 118 - 23771897 CGGGAAUGAGA--CGACACUAGGUGAAAUCCGGUGGCUUGUACUUCGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCUUGGC .(((((..(((--((.((((.((......))))))...........(((((((((..((((((.........))))))))))))))).)))).....)..)))))......(((...))) ( -40.40) >DroVir_CAF1 221424 106 - 1 --------------GGCGCUAGGUAAAGUCCGGCGGCUUGUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUCGA --------------(((((((((...((..(((((...((....(((((((((((..((((((.........))))))))))))))).)).))..)))))..)).)))))))))...... ( -44.70) >DroGri_CAF1 207752 117 - 1 -CGGAAUUUCC--CGGGAUUAGGUAAAAUCGGGCGGUUUAUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAAAUGUUGUUCUCCUUGGCGCCCCCCUU -.((......)--)(((....(((...)))((((............(((((((((..((((((.........)))))))))))))))..(((((............)))))))))))).. ( -39.80) >DroWil_CAF1 165574 120 - 1 GAGGAUUUAGGUUAGACGCUAGGUAAAAUCUGGCGGCUUGUACUUGGGCGGACGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUAGU ((((...(((((.((.((((((((...)))))))).))...)))))(((((.(((..((((((.........))))))))).))))).(((((..............))))).))))... ( -37.84) >DroMoj_CAF1 245049 106 - 1 --------------UAUGCUAGGUAAAAUCCGGCGGCUUGUAUUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUAUUGUUCUCCUUGGCGCCCUUCGA --------------...(((.((......)))))(((.........(((((((((..((((((.........)))))))))))))))..((((..............)))))))...... ( -34.84) >DroAna_CAF1 196806 118 - 1 GGGAAGGUACA--CGCCGCUAGGGGAAGUCCGGCGGCUUGUACUUCGGCGGUCGUUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGUGCCCUUGGC (((.(((((((--.((((((.(((....))))))))).))))))).(((((((((..((((((.........))))))))))))))).........................)))..... ( -48.00) >consensus __GGA_UUA____CGACGCUAGGUAAAAUCCGGCGGCUUGUACUUGGGCGGUCGCUUAAUUGGCUUCUAUAACCAAUUGCGGCCGCCACGUCAUAUGUUGUUCUCCUUGGCGCCCUUCGA .............................((((.(((.........(((((((((..((((((.........)))))))))))))))..((((..............))))))).)))). (-30.48 = -30.09 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:09 2006