
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,600,931 – 4,601,023 |
| Length | 92 |
| Max. P | 0.997974 |

| Location | 4,600,931 – 4,601,023 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.14 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4600931 92 + 23771897 UUCUCUCAGUGCUCACACAUAUCCCAUCGCUUCAGGUUCCAUUUGCGAACGGGUGUGUGUUUCCAAAUGGAUCCUCAUUCUCAUCCACAUGG-- ..............((((((((((..((((....(....)....))))..))))))))))..(((..(((((..........)))))..)))-- ( -23.70) >DroSec_CAF1 177519 94 + 1 UUCUCUCAGUGCUCACACAUAUCCCAUCGCUUCAGUUUCCAUUUGCGAACGGGUGUGUGUUUCCAAGUGGAUCCUCAUCCUCAUCCACAUGGAG ..............((((((((((..((((..............))))..)))))))))).((((.((((((..........)))))).)))). ( -29.54) >DroSim_CAF1 178063 94 + 1 UUCUCUCAGUGCUCACACAUAUCCCAGCGCUUCAGUUUCCAUUUGCGAAUGGGUGUGUGUUUCCAAGUGGAUCCUCAUCCUCAUCCACAUGGAG ..............((((((((((...(((..............)))...)))))))))).((((.((((((..........)))))).)))). ( -29.04) >DroEre_CAF1 186193 92 + 1 UUUUUGUAGUGCUCACACAUAUCCCAUCGUUUCAGGUUCCAUCGGAGACUGGGUGUGUGUUUCCGAGUGGAUCCUCAUCCUCAUCCACAUGG-- ..............(((((((.((((..(((((.(((...))).))))))))))))))))..(((.((((((..........)))))).)))-- ( -25.90) >DroYak_CAF1 183439 92 + 1 UUUGUUUAGUGUUCACACAUAUCCCAUCGUUUCAGGUUCCAUUUGCCAAUGGGUGUGUGUUUCCGAGUGGAUCCUCAUCCUCAUCCACAUGG-- ..............(((((((.(((((.......(((.......))).))))))))))))..(((.((((((..........)))))).)))-- ( -22.20) >consensus UUCUCUCAGUGCUCACACAUAUCCCAUCGCUUCAGGUUCCAUUUGCGAAUGGGUGUGUGUUUCCAAGUGGAUCCUCAUCCUCAUCCACAUGG__ ..............((((((((((..((((..............))))..))))))))))..(((.((((((..........)))))).))).. (-22.82 = -23.14 + 0.32)

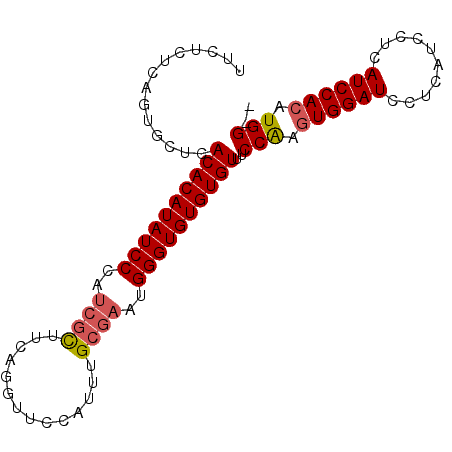

| Location | 4,600,931 – 4,601,023 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -24.10 |
| Energy contribution | -25.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4600931 92 - 23771897 --CCAUGUGGAUGAGAAUGAGGAUCCAUUUGGAAACACACACCCGUUCGCAAAUGGAACCUGAAGCGAUGGGAUAUGUGUGAGCACUGAGAGAA --(((.((((((..........)))))).)))...(((((((((((.(((....(....)....))))))))...))))))............. ( -27.40) >DroSec_CAF1 177519 94 - 1 CUCCAUGUGGAUGAGGAUGAGGAUCCACUUGGAAACACACACCCGUUCGCAAAUGGAAACUGAAGCGAUGGGAUAUGUGUGAGCACUGAGAGAA .((((.((((((..........)))))).))))..(((((((((((.(((....(....)....))))))))...))))))............. ( -33.00) >DroSim_CAF1 178063 94 - 1 CUCCAUGUGGAUGAGGAUGAGGAUCCACUUGGAAACACACACCCAUUCGCAAAUGGAAACUGAAGCGCUGGGAUAUGUGUGAGCACUGAGAGAA .((((.((((((..........)))))).))))..((((((((((..(((....(....)....))).))))...))))))............. ( -30.70) >DroEre_CAF1 186193 92 - 1 --CCAUGUGGAUGAGGAUGAGGAUCCACUCGGAAACACACACCCAGUCUCCGAUGGAACCUGAAACGAUGGGAUAUGUGUGAGCACUACAAAAA --...(((((....(..(.(((.((((.(((((.((.........)).))))))))).))).)..).........(((....)))))))).... ( -22.80) >DroYak_CAF1 183439 92 - 1 --CCAUGUGGAUGAGGAUGAGGAUCCACUCGGAAACACACACCCAUUGGCAAAUGGAACCUGAAACGAUGGGAUAUGUGUGAACACUAAACAAA --((..((((((..........))))))..))...(((((((((((((......(....).....)))))))...))))))............. ( -26.00) >consensus __CCAUGUGGAUGAGGAUGAGGAUCCACUUGGAAACACACACCCAUUCGCAAAUGGAACCUGAAGCGAUGGGAUAUGUGUGAGCACUGAGAGAA ..(((.((((((..........)))))).)))...((((((((((.((((....(....)....))))))))...))))))............. (-24.10 = -25.10 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:05 2006