
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,577,561 – 4,577,717 |
| Length | 156 |
| Max. P | 0.991590 |

| Location | 4,577,561 – 4,577,664 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -22.76 |
| Energy contribution | -22.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4577561 103 - 23771897 GCAUGGGCAUGUGGUGAUGAUGAAAGGUGGGGUUCGGCGAACCAGAAGGGUCAAAGGAGGAAGGGCGGGCGUUGCAAUGGAGGGCAGCGAGCGGCCAAGGCAA ((...(((.(.((((.....((((........))))....)))).)...)))..........((.((..((((((........))))))..)).))...)).. ( -27.60) >DroSec_CAF1 154413 86 - 1 GCAUGGGCAUGUGGGGAUGAUG-----------------AACCAGAAGGGUCAAAGGAGGAAGGGCGGGCGUUGCAAUGGAGGGCAGCGAGCGGCCAAGGCAA ((...(((.(.(((........-----------------..))).)...)))..........((.((..((((((........))))))..)).))...)).. ( -24.00) >DroSim_CAF1 154686 86 - 1 GCAUGGGCAUGUGGGGAUGAUG-----------------AACCAGAAGGGUCAAAGGAGGAAGGGCGGGCGUUGCAAUGGAGGGCAGCGAGCGGCCAAGGCAA ((...(((.(.(((........-----------------..))).)...)))..........((.((..((((((........))))))..)).))...)).. ( -24.00) >consensus GCAUGGGCAUGUGGGGAUGAUG_________________AACCAGAAGGGUCAAAGGAGGAAGGGCGGGCGUUGCAAUGGAGGGCAGCGAGCGGCCAAGGCAA ((...(((.(.(((...........................))).)...)))..........((.((..((((((........))))))..)).))...)).. (-22.76 = -22.76 + -0.00)

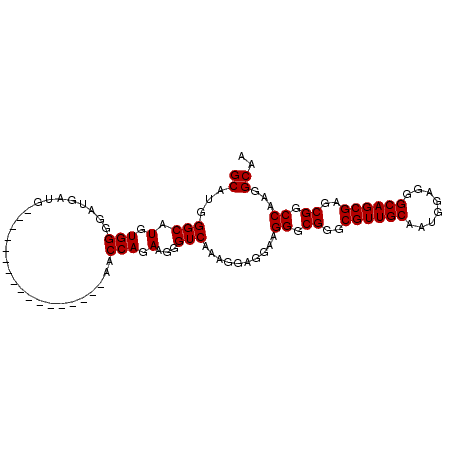

| Location | 4,577,624 – 4,577,717 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4577624 93 + 23771897 UCGCCGAACCCCACCUUUCAUCAUCACCACAUGCCCAUGCCAAAUGCCGAAUGCAAAGUUCGUGCUGACAAAGUCGGCACAAAUCACUGGCCA ..(((((((....................(((....))).....(((.....)))..))))((((((((...))))))))........))).. ( -21.90) >DroSec_CAF1 154476 76 + 1 U-----------------CAUCAUCCCCACAUGCCCAUGCCAAAUGCCGAAUGCAAAAUUCGUGCUAACAAAGUCGGCACAAAUCACUGGCCA .-----------------....................((((......((((.....))))((((..((...))..)))).......)))).. ( -14.90) >DroSim_CAF1 154749 76 + 1 U-----------------CAUCAUCCCCACAUGCCCAUGCCAAAUGCCGAAUGCAAAGUUCGUGCUGACAAAGUCGGCACAAAUCACUGGCCA .-----------------....................((((......((((.....))))((((((((...)))))))).......)))).. ( -19.40) >DroEre_CAF1 162661 88 + 1 UCGCCGAA-CCC--CCAUCACCAUCACCAC--GCCCAGGCCAAAUGCCGAAUGCAAAGUUCGUGCUGACAAAGUCGGCACAAAUCACUGGCCA ..((((((-(..--................--((....))....(((.....)))..))))((((((((...))))))))........))).. ( -24.00) >DroYak_CAF1 159520 90 + 1 UCACCGAAUCCC--CCUUCAUCAUCACCAAA-ACCCAUGCGAAAUGCCGAAUGCAAAGUUCGUGCUGACAAAGUCGGCACAAAUCACUGGCCA .....(((....--..)))............-.............(((((((.....))))((((((((...))))))))........))).. ( -18.80) >consensus UC_CCGAA_CCC__C__UCAUCAUCACCACAUGCCCAUGCCAAAUGCCGAAUGCAAAGUUCGUGCUGACAAAGUCGGCACAAAUCACUGGCCA .............................................(((((((.....))))((((((((...))))))))........))).. (-17.86 = -17.90 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:56 2006