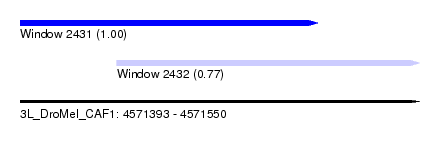
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,571,393 – 4,571,550 |
| Length | 157 |
| Max. P | 0.999907 |

| Location | 4,571,393 – 4,571,510 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -26.50 |
| Energy contribution | -26.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4571393 117 + 23771897 UAUUGCUU--CAUUUUCAAAGGCUGGGCAACUCUGACGACCAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUCCCCUGAGAUGCACAUUUU-GAGUUACUAUCCUUUAGGAAA ........--..(((..(((((.(((..(((((((.....)).....(((((((((((((((.....................)))))))))))))))-))))).))).)))))..))). ( -30.70) >DroSec_CAF1 148298 117 + 1 UAUUGCUU--CAUUUUCAAAGGCUGGGUAACUCUGACGACCAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUUCCCUGAGCUGCACAUUUU-GAGUUACUAUCCUUUAGGAAA ........--..(((..(((((...((((((((((.....)).....((((((((((.((((.....................)))).))))))))))-))))))))..)))))..))). ( -29.20) >DroSim_CAF1 148568 117 + 1 UAUUGCUU--CAUUUUCAAAGGCUGGUUAACUCUGACGACCAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUUCCCUGAGCUGCACGUUUU-GAGUUACUUACCUUUAGGAAA ........--..(((..(((((..(((.(((((((.....)).....((((((((((.((((.....................)))).))))))))))-))))))))..)))))..))). ( -25.90) >DroEre_CAF1 156377 119 + 1 UAUUGCUUCUCAUUUUCAAAGGCUAGUUGGCUCUGACGACCAGCUUAAAAAUGUGUAUCUCAAAUUCCCCUAUCCCUCCC-CCCGAGAUGCACAUUUUAGAGUUAGUUUCCUUUUGGGAA ....(((.(((........(((((.((((.......)))).))))).((((((((((((((...................-...)))))))))))))).)))..)))((((....)))). ( -32.55) >DroYak_CAF1 153245 119 + 1 UAUUGCUUCGCAUUUUCAAAGGCUGGGUAACUCUGACGUCCAGCUUAAAAAUAUGUAUCUCAAAU-CCCAUAGCCCUCCCUCCCGAGAUGCACAUUUUAGAGUUACUUUCCUUUAGGGAA ...(((...)))((..((((((..((((((((((...(.....)...(((((.((((((((....-..................)))))))).))))))))))))))).))))).)..)) ( -27.75) >consensus UAUUGCUU__CAUUUUCAAAGGCUGGGUAACUCUGACGACCAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUCCCCUGAGAUGCACAUUUU_GAGUUACUUUCCUUUAGGAAA ............((((((((((...((((((((((.....)).....((((((((((((((.......................)))))))))))))).))))))))..))))).))))) (-26.50 = -26.54 + 0.04)

| Location | 4,571,431 – 4,571,550 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4571431 119 + 23771897 CAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUCCCCUGAGAUGCACAUUUU-GAGUUACUAUCCUUUAGGAAAUGCUGGAAAGCUGGCUAUGUAAUUCUUGUUAGGGAAUGUA ...(((((((((((((((((((.....................)))))))))))))))-(((((((..(((....)))...((..(....)..))...)))))))....))))....... ( -32.70) >DroSec_CAF1 148336 115 + 1 CAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUUCCCUGAGCUGCACAUUUU-GAGUUACUAUCCUUUAGGAAAUG----AAAGCUGGCUAUGUAAUUUUUGUUAGGGAAUGUA .(((((((((.((((((.((((.....................)))).))))))))))-)))))....(((((..((((((.----..((....)).....))))))...)))))..... ( -23.70) >DroSim_CAF1 148606 115 + 1 CAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUUCCCUGAGCUGCACGUUUU-GAGUUACUUACCUUUAGGAAAUG----AAAGCUGGCUAUGCAAUUUUUGUUAGGGCAUGUA .(((((((((.((((((.((((.....................)))).))))))))))-))))).....(((...((((((.----...((.......)).))))))...)))....... ( -23.10) >DroEre_CAF1 156417 115 + 1 CAGCUUAAAAAUGUGUAUCUCAAAUUCCCCUAUCCCUCCC-CCCGAGAUGCACAUUUUAGAGUUAGUUUCCUUUUGGGAAUG----AAAGCUGGAUAUGUAUUUUUUGUUAGGGAAUGUU ((((((.((((((((((((((...................-...)))))))))))))).......((((((....)))))).----.))))))......(((((((.....))))))).. ( -30.05) >DroYak_CAF1 153285 111 + 1 CAGCUUAAAAAUAUGUAUCUCAAAU-CCCAUAGCCCUCCCUCCCGAGAUGCACAUUUUAGAGUUACUUUCCUUUAGGGAAUG----AAAGCUGGAUAUGCAUUUUUUGUUAGGGAA---- ..(((........((.....))...-.....)))..(((((..(((((((((...(((((..((...((((.....))))..----))..)))))..))))))..)))..))))).---- ( -24.79) >consensus CAACUUAAAAAUGUGUAUCUCAAAUCCCCAUAUCCCUUUCCCCUGAGAUGCACAUUUU_GAGUUACUUUCCUUUAGGAAAUG____AAAGCUGGCUAUGUAAUUUUUGUUAGGGAAUGUA .(((((.((((((((((((((.......................)))))))))))))).)))))....(((((..((((((........((.......)).))))))...)))))..... (-21.60 = -22.20 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:51 2006