
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,561,415 – 4,561,535 |
| Length | 120 |
| Max. P | 0.997554 |

| Location | 4,561,415 – 4,561,512 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -47.62 |
| Consensus MFE | -42.03 |
| Energy contribution | -43.00 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.12 |
| Mean z-score | -5.12 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
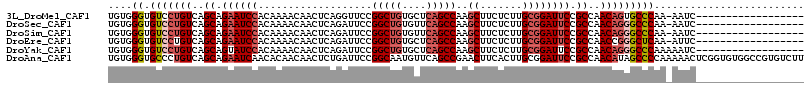
>3L_DroMel_CAF1 4561415 97 + 23771897 ------------------GAUU-UUGGGCACUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAACCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACA ------------------....-.((((..((((..(((((((((((((((..((((((((((....)))))(....).))))).)))))))))))))))..))))....)))).. ( -49.60) >DroSec_CAF1 138402 97 + 1 ------------------GAUU-UUGGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAACACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACA ------------------....-.((((.(((((..(((((((((((((((..((((((((((....))))).......))))).)))))))))))))))..)))))...)))).. ( -50.71) >DroSim_CAF1 138632 97 + 1 ------------------GAUU-UUGGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAACACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACA ------------------....-.((((.(((((..(((((((((((((((..((((((((((....))))).......))))).)))))))))))))))..)))))...)))).. ( -50.71) >DroEre_CAF1 146461 97 + 1 ------------------GAAU-UUGAGCCCGGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACA ------------------....-....(.((.((..(((((((((((((((..((((((((((....))))).......))))).)))))))))))))))..)).)).)....... ( -41.41) >DroYak_CAF1 142932 98 + 1 ------------------GAUUUUUGGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAAUCUGAGUUGUUUUGUGGAUACUGCUGACAGGACACCCACA ------------------......((((.(((((..((((.((((((((((..((((((((((....))))).......))))).)))))))))).))))..)))))...)))).. ( -46.31) >DroAna_CAF1 140008 116 + 1 AAGACACGGCCACACCGAGUUUUUGGGGCUAUGUUGGCGGAAUCCGCAAGUGAAGUUCGGCUGAACAUUGCCGGAAUCAGAGUUGUUGUGUUGAUUCUGCUGACAGGGCACCCACA (((((.(((.....))).)))))(((((((.(((..((((((((.(((((..(..((((((........)))))).......)..)).))).))))))))..))).))).)))).. ( -47.00) >consensus __________________GAUU_UUGGGCCCUGUUGGCGGAAUCCGCAAGAGAAGCUUGGCUGAACACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACA .........................(((.(((((..(((((((((((((((..((((((((((....)))))(....).))))).)))))))))))))))..)))))...)))... (-42.03 = -43.00 + 0.97)


| Location | 4,561,415 – 4,561,512 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -29.59 |
| Energy contribution | -30.82 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4561415 97 - 23771897 UGUGGGUGUCCUGUCAGCAGAAUCCACAAAACAACUCAGGUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGUGCCCAA-AAUC------------------ ..(((((...((((..((.((((((...................(((((....)))))..((.......)))))))).))..)))).))))).-....------------------ ( -33.90) >DroSec_CAF1 138402 97 - 1 UGUGGGUGUCCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGUUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCCAA-AAUC------------------ ....((.(((((((..((.((((((...................(((((....)))))..((.......)))))))).))..)))))))))..-....------------------ ( -37.40) >DroSim_CAF1 138632 97 - 1 UGUGGGUGUCCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGUUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCCAA-AAUC------------------ ....((.(((((((..((.((((((...................(((((....)))))..((.......)))))))).))..)))))))))..-....------------------ ( -37.40) >DroEre_CAF1 146461 97 - 1 UGUGGGUGUCCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACCGGGCUCAA-AUUC------------------ (((((((...(((....))).)))))))..........((.((((((((....)))))..((.......))))).)).(((.....)))....-....------------------ ( -30.90) >DroYak_CAF1 142932 98 - 1 UGUGGGUGUCCUGUCAGCAGUAUCCACAAAACAACUCAGAUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCCAAAAAUC------------------ ....((.(((((((..((.(.((((...................(((((....)))))..((.......)))))).).))..))))))))).......------------------ ( -33.00) >DroAna_CAF1 140008 116 - 1 UGUGGGUGCCCUGUCAGCAGAAUCAACACAACAACUCUGAUUCCGGCAAUGUUCAGCCGAACUUCACUUGCGGAUUCCGCCAACAUAGCCCCAAAAACUCGGUGUGGCCGUGUCUU ..((((.((..(((..((.(((((..((...((....))....((((........)))).........))..))))).))..)))..))))))...((.(((.....))).))... ( -31.20) >consensus UGUGGGUGUCCUGUCAGCAGAAUCCACAAAACAACUCAGAUUCCGGCUGUGCUCAGCCAAGCUUCUCUUGCGGAUUCCGCCAACAGGGCCCAA_AAUC__________________ ....((.(((((((..((.((((((...................(((((....)))))..((.......)))))))).))..)))))))))......................... (-29.59 = -30.82 + 1.22)


| Location | 4,561,436 – 4,561,535 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 97.98 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -30.27 |
| Energy contribution | -30.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
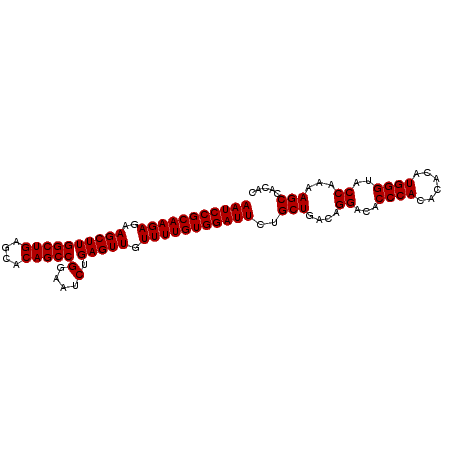
>3L_DroMel_CAF1 4561436 99 + 23771897 AAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAACCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACACACAUGGGUACCAAAAGCCACAC (((((((((((..((((((((((....)))))(....).))))).)))))))))))..(((....((..(((((......))))).))...)))..... ( -32.90) >DroSim_CAF1 138653 99 + 1 AAUCCGCAAGAGAAGCUUGGCUGAACACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACACACAUGGGUACCAAAAGCCACAC (((((((((((..((((((((((....))))).......))))).)))))))))))..(((....((..(((((......))))).))...)))..... ( -31.51) >DroEre_CAF1 146482 99 + 1 AAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACACACAUGGGCACCAAAAGCCACAC (((((((((((..((((((((((....))))).......))))).)))))))))))..(((....((...((((......))))..))...)))..... ( -30.01) >consensus AAUCCGCAAGAGAAGCUUGGCUGAGCACAGCCGGAAUCUGAGUUGUUUUGUGGAUUCUGCUGACAGGACACCCACACACAUGGGUACCAAAAGCCACAC (((((((((((..((((((((((....)))))(....).))))).)))))))))))..(((....((...((((......))))..))...)))..... (-30.27 = -30.27 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:46 2006