
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,539,401 – 4,539,507 |
| Length | 106 |
| Max. P | 0.959508 |

| Location | 4,539,401 – 4,539,507 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -29.76 |
| Energy contribution | -29.64 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
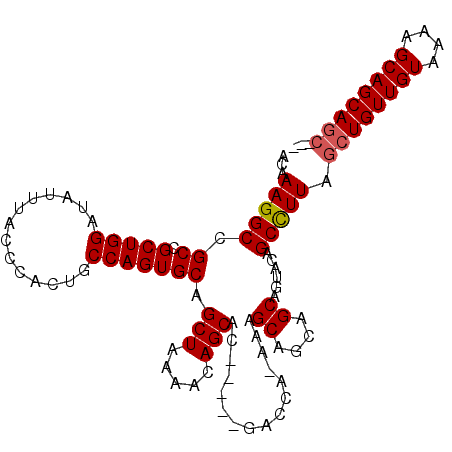
>3L_DroMel_CAF1 4539401 106 + 23771897 ---GCUGCUGCUUUUACAACAGCUAAGGCUAUACUGCUGCUGCUUUUUGGUCGGUUGGUGCUGUUUUAGCUGCACUGGCAGUGGGUAAAUAUCCAGCGGCGGCCUUUGU ---((((((((((((((..((((((((((..((((...((((((....)).)))).))))..))))))))))(((.....))).))))).....)))))))))...... ( -39.70) >DroSec_CAF1 116721 100 + 1 ---GCUGCUGCUUUUACAACAGCUAAGGCUGUACUGCUGCUGCUUU-UGGUC-----GUGCUGUUUUAGCUGCACUGGCAGUGGGUAAAUAUCCAGCGGCGGCCUUUGU ---((((((((((((((.(((((....))))).(..(((((((...-..)).-----((((.((....)).)))).)))))..)))))).....)))))))))...... ( -40.50) >DroEre_CAF1 122598 97 + 1 -----UGCUGCUUUUACAACAGCUAAAGCUGAACUGCUGCUGCUUU-G-GUC-----CUGCUGUGUUAGCUGCACUGGCAGUGGGUAAAUAUCCAGCGGCGGCCUUUGU -----..........((((((((....))))....((((((((...-.-...-----(((((((((.....)))).)))))(((((....)))))))))))))..)))) ( -34.50) >DroYak_CAF1 120156 103 + 1 GUUGCUGCUGCUUUUACAACAGCUAAAGCUAUACUGCUCCAGCUUU-GGGUC-----UUGCUGGCUUAGCUGCACUGGCAGUGGGUAAAUAUCCAGCGGCGGCCUUUGU ((((((((((..(((((...(((....))).(((((((.(((((..-(((((-----.....))))))))))....))))))).)))))....))))))))))...... ( -39.60) >consensus ___GCUGCUGCUUUUACAACAGCUAAAGCUAUACUGCUGCUGCUUU_GGGUC_____GUGCUGUUUUAGCUGCACUGGCAGUGGGUAAAUAUCCAGCGGCGGCCUUUGU ...((((((((((((((..((((((((((......)))((.((................)).)).)))))))(((.....))).))))).....)))))))))...... (-29.76 = -29.64 + -0.12)



| Location | 4,539,401 – 4,539,507 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4539401 106 - 23771897 ACAAAGGCCGCCGCUGGAUAUUUACCCACUGCCAGUGCAGCUAAAACAGCACCAACCGACCAAAAAGCAGCAGCAGUAUAGCCUUAGCUGUUGUAAAAGCAGCAGC--- ...(((((.......((........))(((((...(((.(((.....)))..(....)........)))...)))))...))))).((((((((....))))))))--- ( -27.70) >DroSec_CAF1 116721 100 - 1 ACAAAGGCCGCCGCUGGAUAUUUACCCACUGCCAGUGCAGCUAAAACAGCAC-----GACCA-AAAGCAGCAGCAGUACAGCCUUAGCUGUUGUAAAAGCAGCAGC--- ...(((((.((.(.(((........)))).))..((((.(((.....)))..-----.....-...((....)).)))).))))).((((((((....))))))))--- ( -29.00) >DroEre_CAF1 122598 97 - 1 ACAAAGGCCGCCGCUGGAUAUUUACCCACUGCCAGUGCAGCUAACACAGCAG-----GAC-C-AAAGCAGCAGCAGUUCAGCUUUAGCUGUUGUAAAAGCAGCA----- .....((((((.(((((..............))))))).(((.....))).)-----).)-)-((((((((....)))..))))).((((((.....)))))).----- ( -26.84) >DroYak_CAF1 120156 103 - 1 ACAAAGGCCGCCGCUGGAUAUUUACCCACUGCCAGUGCAGCUAAGCCAGCAA-----GACCC-AAAGCUGGAGCAGUAUAGCUUUAGCUGUUGUAAAAGCAGCAGCAAC ...(((((.((.(((((..............))))))).(((...(((((..-----.....-...))))))))......))))).((((((((....))))))))... ( -33.04) >consensus ACAAAGGCCGCCGCUGGAUAUUUACCCACUGCCAGUGCAGCUAAAACAGCAC_____GACCA_AAAGCAGCAGCAGUACAGCCUUAGCUGUUGUAAAAGCAGCAGC___ ...(((((.((.(((((..............))))))).(((.....)))................((....))......))))).((((((((....))))))))... (-24.67 = -24.92 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:31 2006