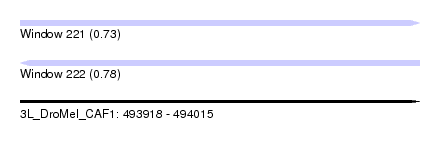
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 493,918 – 494,015 |
| Length | 97 |
| Max. P | 0.776642 |

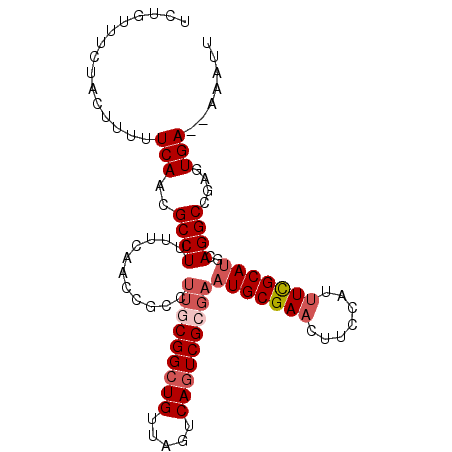
| Location | 493,918 – 494,015 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.56 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -17.66 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
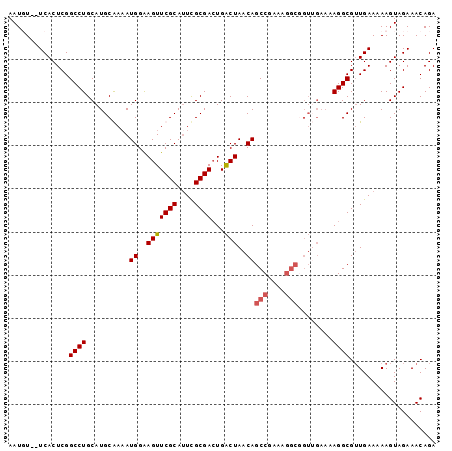
>3L_DroMel_CAF1 493918 97 + 23771897 AAUUU--UCACUCGGCCUGCAUGCAAAAUGGAAGUUCGCAUUCGCGACUGACUAACAGCCGAAAGGCAGUGGAAAAGGCGUUGAAAAAGUAGAAACAGA ..(((--(((..((.(((...(((....((..(((((((....))))...)))..))(((....))).)))....))))).))))))............ ( -25.70) >DroSec_CAF1 74235 96 + 1 AAUGU--UCACUCGGCCUGCAUGCGA-AUGGAAGUUCGCAUUCGCGACUGACUAACAGCCGCAAGGCGGUUGAAAAGGCGGUGAAAAAGUAGAAACAGA ....(--(((((..((((((((((((-((....))))))))..)).......((((.(((....))).))))...)))))))))).............. ( -32.70) >DroSim_CAF1 75572 97 + 1 AAUGU--UCACUCGGCCUGCAUGCGAAAUGGAAGUUCGCAUUCGCGACUGACUAACAGCCGCAAGGCGGUUGAAAAGGCGUUGAAAAAGUAGAAACAGA (((((--(...((((((((((((((((.......)))))))..)))...........(((....)))))))))...))))))................. ( -27.30) >DroYak_CAF1 78100 87 + 1 AAUUUUAUCACUCGGCCUGCAUGCAAAAUGGGAGUUCGCAAUUGCGAAUGGCUAACAGCCGA------------AAGGCGUUGAAAAAGUAGAAACAGA .(((((.(((...((((..(((.....)))...((((((....))))))))))....(((..------------..)))..))).)))))......... ( -24.50) >consensus AAUGU__UCACUCGGCCUGCAUGCAAAAUGGAAGUUCGCAUUCGCGACUGACUAACAGCCGAAAGGCGGUUGAAAAGGCGUUGAAAAAGUAGAAACAGA ..............((((..........((..(((((((....))))...)))..))(((....)))........)))).................... (-17.66 = -18.23 + 0.56)

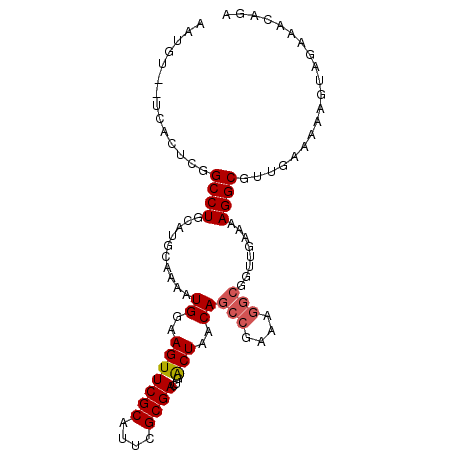
| Location | 493,918 – 494,015 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.56 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.60 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 493918 97 - 23771897 UCUGUUUCUACUUUUUCAACGCCUUUUCCACUGCCUUUCGGCUGUUAGUCAGUCGCGAAUGCGAACUUCCAUUUUGCAUGCAGGCCGAGUGA--AAAUU ............((((((..((((...............(((((.....)))))((..(((((((.......)))))))))))))....)))--))).. ( -21.70) >DroSec_CAF1 74235 96 - 1 UCUGUUUCUACUUUUUCACCGCCUUUUCAACCGCCUUGCGGCUGUUAGUCAGUCGCGAAUGCGAACUUCCAU-UCGCAUGCAGGCCGAGUGA--ACAUU ..............((((((((((...........(((((((((.....)))))))))(((((((......)-))))))..))).)).))))--).... ( -30.50) >DroSim_CAF1 75572 97 - 1 UCUGUUUCUACUUUUUCAACGCCUUUUCAACCGCCUUGCGGCUGUUAGUCAGUCGCGAAUGCGAACUUCCAUUUCGCAUGCAGGCCGAGUGA--ACAUU .........................((((..(((((((((((((.....)))))))))(((((((.......)))))))...)).))..)))--).... ( -27.20) >DroYak_CAF1 78100 87 - 1 UCUGUUUCUACUUUUUCAACGCCUU------------UCGGCUGUUAGCCAUUCGCAAUUGCGAACUCCCAUUUUGCAUGCAGGCCGAGUGAUAAAAUU ...........(((.(((..((((.------------..(((.....)))....(((..((((((.......)))))))))))))....))).)))... ( -18.60) >consensus UCUGUUUCUACUUUUUCAACGCCUUUUCAACCGCCUUGCGGCUGUUAGUCAGUCGCGAAUGCGAACUUCCAUUUCGCAUGCAGGCCGAGUGA__AAAUU ...............(((..((((...........(((((((((.....)))))))))(((((((.......)))))))..))))....)))....... (-18.10 = -19.60 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:01 2006