| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,522,829 – 4,522,948 |
| Length | 119 |
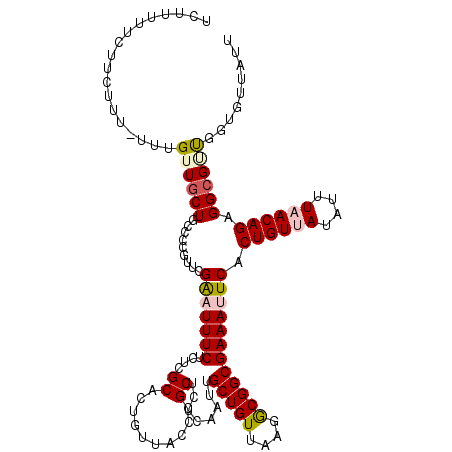
| Max. P | 0.593494 |

| Location | 4,522,829 – 4,522,948 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -17.31 |
| Energy contribution | -18.38 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4522829 119 + 23771897 ACUAACAACAACGCCUCUGUUAAAUAUAACAGUGAAUUUCGCCGCUUUAACAGCAACUGAGAGCGGCUAACAGUGCGAGAGAAAUUCGAACGGGGCAGCAACAAA-AAAGAAGAAAAAGA ............((((((((((....))))).((((((((((((((((..(((...)))))))))))...(.....)...))))))))...))))).........-.............. ( -29.30) >DroSec_CAF1 100473 116 + 1 ---AACUACAACGCCUCUGUUAAAUAUAACAGUGAGUUUCGCCGCCUUAACAGCAACUGAGAGCGGGUAACAGCGCGAGAGAAAUUCGAACG-GGCAGCAACAAAAAAAGAAGAAAAAGA ---.........((((((((((....))))))((((((((.((((((((........)))).)))).......(....).))))))))...)-)))........................ ( -27.70) >DroEre_CAF1 106606 118 + 1 AAAAAUACCAUCGCCUCUGUGAAACUUAACAGUGAAUUUCGCCGUCUUAACAGCAAUUGAGAGCGGGUAACAGUGCGAGAGAAA-CCGAACGAGCCAGCAACAAA-AAAGAAGAAGAAUA .......(..((((..((((((((.(((....))).)))).(((((((((......)))))).)))...)))).))))..)...-....................-.............. ( -19.40) >DroYak_CAF1 102821 119 + 1 UAUGACACCGCCGCCUCUGUAAAAUAUAACAGUGAAUUUCGCCGCAUUAACAGCAAUUGAGAGCGGCUAACAGUGCGAGAGAAAUUCGAACGGGGCAGUAACAAA-AAAGAAGAAAAAGA ..((..((.(((.((.((((........))))((((((((.(((((((...(((...(....)..)))...)))))).).))))))))...))))).))..))..-.............. ( -28.70) >consensus AAUAACAACAACGCCUCUGUUAAAUAUAACAGUGAAUUUCGCCGCCUUAACAGCAACUGAGAGCGGCUAACAGUGCGAGAGAAAUUCGAACGGGGCAGCAACAAA_AAAGAAGAAAAAGA ............((((((((((....))))).((((((((.((((.(((...((........))...)))....))).).))))))))...)))))........................ (-17.31 = -18.38 + 1.06)

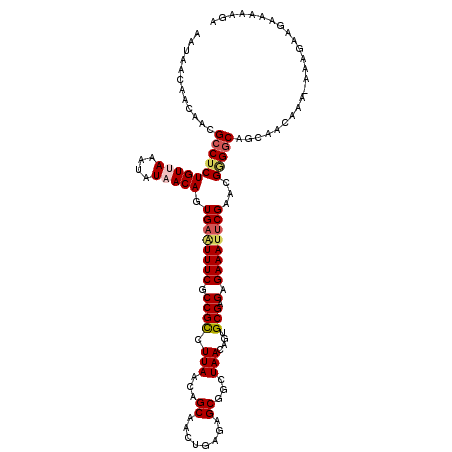
| Location | 4,522,829 – 4,522,948 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -19.38 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4522829 119 - 23771897 UCUUUUUCUUCUUU-UUUGUUGCUGCCCCGUUCGAAUUUCUCUCGCACUGUUAGCCGCUCUCAGUUGCUGUUAAAGCGGCGAAAUUCACUGUUAUAUUUAACAGAGGCGUUGUUGUUAGU ..............-.........(((......(((((((.(.(((....(((((.((........)).))))).)))).))))))).((((((....)))))).)))............ ( -28.20) >DroSec_CAF1 100473 116 - 1 UCUUUUUCUUCUUUUUUUGUUGCUGCC-CGUUCGAAUUUCUCUCGCGCUGUUACCCGCUCUCAGUUGCUGUUAAGGCGGCGAAACUCACUGUUAUAUUUAACAGAGGCGUUGUAGUU--- .....................(((((.-((((.((.((((....(((........))).....((((((.....)))))))))).)).((((((....)))))).))))..))))).--- ( -25.30) >DroEre_CAF1 106606 118 - 1 UAUUCUUCUUCUUU-UUUGUUGCUGGCUCGUUCGG-UUUCUCUCGCACUGUUACCCGCUCUCAAUUGCUGUUAAGACGGCGAAAUUCACUGUUAAGUUUCACAGAGGCGAUGGUAUUUUU ..............-...(..(((((.....))))-)..)...........(((((((.(((....(((((....)))))((((((........))))))...))))))..))))..... ( -22.90) >DroYak_CAF1 102821 119 - 1 UCUUUUUCUUCUUU-UUUGUUACUGCCCCGUUCGAAUUUCUCUCGCACUGUUAGCCGCUCUCAAUUGCUGUUAAUGCGGCGAAAUUCACUGUUAUAUUUUACAGAGGCGGCGGUGUCAUA ..............-..((.(((((((((....(((((((.(.((((...(((((.((........)).)))))))))).))))))).((((........)))).)).))))))).)).. ( -36.50) >consensus UCUUUUUCUUCUUU_UUUGUUGCUGCCCCGUUCGAAUUUCUCUCGCACUGUUACCCGCUCUCAAUUGCUGUUAAGGCGGCGAAAUUCACUGUUAUAUUUAACAGAGGCGUUGGUGUUAUU ..................((((((.........(((((((....((..........))........(((((....)))))))))))).((((((....)))))).))))))......... (-19.38 = -20.50 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:24 2006