
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,463,354 – 4,463,454 |
| Length | 100 |
| Max. P | 0.752315 |

| Location | 4,463,354 – 4,463,454 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -22.26 |
| Energy contribution | -21.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
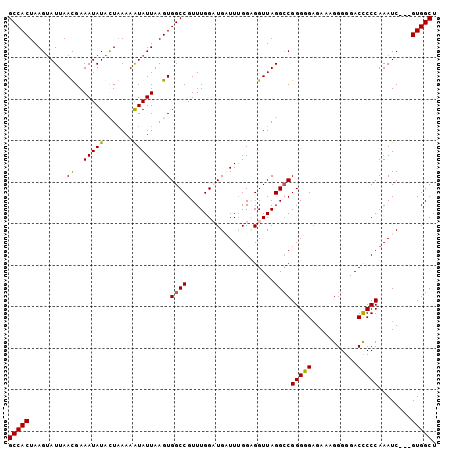
>3L_DroMel_CAF1 4463354 100 + 23771897 GCCACUAAGUAUUAACGAAAUAUACUUAAAAUAUUAAGUGGCCGUUUGGAUGAUUUGGCGGUUAGGCCGGGGGUGAAAGGGGGACUCCCAAAUC---GUGGCU (((((..........((((...(((((((....)))))))....))))...((((((((......)))((((((.........)))))))))))---))))). ( -31.90) >DroSec_CAF1 40899 100 + 1 GCCACUAAGUAUUAACGAAAUAUAUUAAAAAUAUUAAGUGGCCGUUUGGAUGAUUUGGAGGUUAGGCCGGGGGAGAAAGGGGGACCCCCAAAUC---GUGGCU ((((((.((((((................)))))).)))))).......(((((((((.((((...((..........))..)))).)))))))---)).... ( -28.69) >DroEre_CAF1 44584 100 + 1 GCCACUAAGAAUUAAUGAAAUAUACUAAAAAUAUUAAGUGGCCGUUUGGCUGCUUUGGAGGUUAGGACGGGGGAGAAAGGGGGACUCCCAAAUC---GUGGCU (((((...................((((.......(((..(((....)))..)))......))))((...(((((.........)))))...))---))))). ( -30.22) >DroYak_CAF1 42773 103 + 1 GCCACUAAGUAUUAAUGAAAUAUAUUAAAAGUAUUAAGUGGCCGUUUGGCUGUUUAGGUGGUUAGGCCGGGGGAGAAAGGGGGACCCCCAAAUCGUCGUGGCU ((((((..(((((.....)))))...........((((..(((....)))..))))))))))..(((((.((..((...(((....)))...)).)).))))) ( -32.00) >consensus GCCACUAAGUAUUAACGAAAUAUACUAAAAAUAUUAAGUGGCCGUUUGGAUGAUUUGGAGGUUAGGCCGGGGGAGAAAGGGGGACCCCCAAAUC___GUGGCU (((((.........((..(((((.......)))))..))((((.....................))))(((((...........)))))........))))). (-22.26 = -21.83 + -0.44)


| Location | 4,463,354 – 4,463,454 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
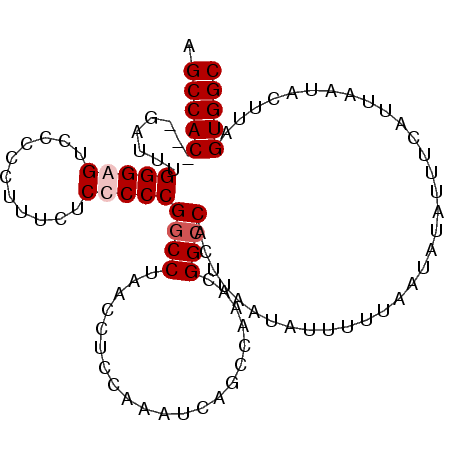
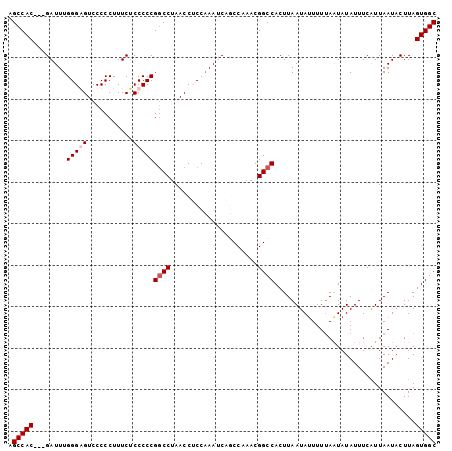
>3L_DroMel_CAF1 4463354 100 - 23771897 AGCCAC---GAUUUGGGAGUCCCCCUUUCACCCCCGGCCUAACCGCCAAAUCAUCCAAACGGCCACUUAAUAUUUUAAGUAUAUUUCGUUAAUACUUAGUGGC .(((((---((((((((((.......))).....(((.....)))))))))).....(((((..((((((....)))))).....)))))........))))) ( -25.20) >DroSec_CAF1 40899 100 - 1 AGCCAC---GAUUUGGGGGUCCCCCUUUCUCCCCCGGCCUAACCUCCAAAUCAUCCAAACGGCCACUUAAUAUUUUUAAUAUAUUUCGUUAAUACUUAGUGGC .(((((---(((((((((((...((..........))....))))))))))).....(((((......(((((.......))))))))))........))))) ( -26.50) >DroEre_CAF1 44584 100 - 1 AGCCAC---GAUUUGGGAGUCCCCCUUUCUCCCCCGUCCUAACCUCCAAAGCAGCCAAACGGCCACUUAAUAUUUUUAGUAUAUUUCAUUAAUUCUUAGUGGC .(((((---(((..(((((.........)))))..)))((((......(((..(((....)))..))).......))))...................))))) ( -22.62) >DroYak_CAF1 42773 103 - 1 AGCCACGACGAUUUGGGGGUCCCCCUUUCUCCCCCGGCCUAACCACCUAAACAGCCAAACGGCCACUUAAUACUUUUAAUAUAUUUCAUUAAUACUUAGUGGC .(((((........(((((...........)))))((((.....................))))..................................))))) ( -24.40) >consensus AGCCAC___GAUUUGGGAGUCCCCCUUUCUCCCCCGGCCUAACCUCCAAAUCAGCCAAACGGCCACUUAAUAUUUUUAAUAUAUUUCAUUAAUACUUAGUGGC .(((((........(((((...........)))))((((.....................))))..................................))))) (-17.50 = -18.25 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:16 2006