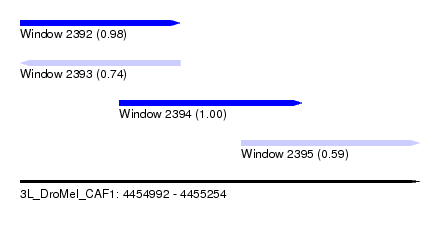
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,454,992 – 4,455,254 |
| Length | 262 |
| Max. P | 0.999606 |

| Location | 4,454,992 – 4,455,097 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -31.41 |
| Energy contribution | -30.73 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
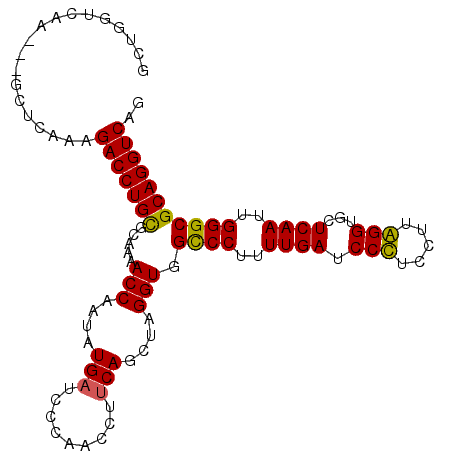
>3L_DroMel_CAF1 4454992 105 + 23771897 GCUGGUCAAAGACCUCAAAGACCUGCGCAAAACCAAUAUGAUCCCAACCUUCAGCUAGGUGGCCCUUUUGACCCCUCCGUGGGUGCUCAAUUGGGCGCAGGUCAG ...((((...)))).....(((((((((...............(((.(((......)))))).((..((((((((.....))).).))))..))))))))))).. ( -40.10) >DroSec_CAF1 32238 102 + 1 GCUGGUCAA---GCUCAAAGACCUGUGCAAAACCAAUAUGAUCCCAAUCUUCAGCUAGGUGGCCCUUUUGAUCCUUCUUUAGGUGCUCAAUUGGGCGCAGGUCAG (((.....)---)).....(((((((.....(((....(((.........)))....))).((((..((((.(((.....)))...))))..))))))))))).. ( -33.10) >DroSim_CAF1 32176 102 + 1 GCUGGUCAA---GCUCAAAGACCUGUGCAAAACCAAUAUGAUCCCAAUCUUCAGCUCGGUGGCCCAUUUGAUCCUUCUUUAGGUGCUCAAUUGGGCGCAGGUCAG (((.....)---)).....(((((((.....(((....(((.........)))....))).(((((.((((.(((.....)))...)))).)))))))))))).. ( -34.10) >DroYak_CAF1 33883 102 + 1 GGAGGUCAA---GCUCAAAGACCUGCGCAAAACCAAUAUGAUCCCAACCUACAGCUAGGUCGUCCUUUUGAUCCCUCCUUGGGUGCUCAAUUGGGCGCAGGUCAG .(((.....---.)))...(((((((((..................(((((....)))))...((..((((.(((.....)))...))))..))))))))))).. ( -36.20) >consensus GCUGGUCAA___GCUCAAAGACCUGCGCAAAACCAAUAUGAUCCCAACCUUCAGCUAGGUGGCCCUUUUGAUCCCUCCUUAGGUGCUCAAUUGGGCGCAGGUCAG ...................(((((((.....(((....(((.........)))....))).((((..((((.(((.....)))...))))..))))))))))).. (-31.41 = -30.73 + -0.69)


| Location | 4,454,992 – 4,455,097 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -31.95 |
| Energy contribution | -31.33 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
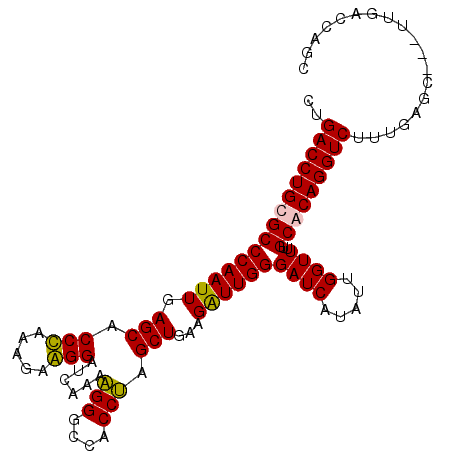
>3L_DroMel_CAF1 4454992 105 - 23771897 CUGACCUGCGCCCAAUUGAGCACCCACGGAGGGGUCAAAAGGGCCACCUAGCUGAAGGUUGGGAUCAUAUUGGUUUUGCGCAGGUCUUUGAGGUCUUUGACCAGC ..(((((((((((((((((...((((((((((((((.....)))).)))..))).....)))).))).)))))....))))))))).....((((...))))... ( -42.10) >DroSec_CAF1 32238 102 - 1 CUGACCUGCGCCCAAUUGAGCACCUAAAGAAGGAUCAAAAGGGCCACCUAGCUGAAGAUUGGGAUCAUAUUGGUUUUGCACAGGUCUUUGAGC---UUGACCAGC ..((((((.((((((((.(((.(((.....)))......(((....))).)))...))))))((((.....))))..)).)))))).....((---(.....))) ( -28.60) >DroSim_CAF1 32176 102 - 1 CUGACCUGCGCCCAAUUGAGCACCUAAAGAAGGAUCAAAUGGGCCACCGAGCUGAAGAUUGGGAUCAUAUUGGUUUUGCACAGGUCUUUGAGC---UUGACCAGC (((...((.(((((.((((...(((.....))).)))).))))))).(((((((((((((..((((.....))))..).....)))))).)))---)))..))). ( -30.90) >DroYak_CAF1 33883 102 - 1 CUGACCUGCGCCCAAUUGAGCACCCAAGGAGGGAUCAAAAGGACGACCUAGCUGUAGGUUGGGAUCAUAUUGGUUUUGCGCAGGUCUUUGAGC---UUGACCUCC ..(((((((((((((((((.(.(((.....)))..........(((((((....))))))).).))).)))))....)))))))))...(((.---.....))). ( -36.60) >consensus CUGACCUGCGCCCAAUUGAGCACCCAAAGAAGGAUCAAAAGGGCCACCUAGCUGAAGAUUGGGAUCAUAUUGGUUUUGCACAGGUCUUUGAGC___UUGACCAGC ..(((((((((((((((.(((.(((.....)))......(((....))).)))...))))))((((.....))))..)))))))))................... (-31.95 = -31.33 + -0.63)

| Location | 4,455,057 – 4,455,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.67 |
| Mean single sequence MFE | -47.54 |
| Consensus MFE | -33.34 |
| Energy contribution | -36.70 |
| Covariance contribution | 3.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4455057 120 + 23771897 UUUUGACCCCUCCGUGGGUGCUCAAUUGGGCGCAGGUCAGGGUGUUUCUAACCAAAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCUACAGGUGCUCAAUUGGGCGCAGGUGAGGGUGU .....(((((.((....((((((((((((((((.((((..(((.......)))...))))..(((((..((((........)))))))))..)))))))))))))))).)).).)))).. ( -56.10) >DroPse_CAF1 39689 92 + 1 -------------------GCACAAUUUGG---AGGUCAGCUGCCCGCUGCCCAACAGCCGAAUCAACUCGGA------AAUCCCGCUCUGGGCGCUCAGCUGGGAGUUGGACAGGCUCC -------------------((.....(((.---.((.((((.....)))).))..)))((((......)))).------..(((.(((((.(((.....))).))))).)))...))... ( -31.80) >DroSec_CAF1 32300 120 + 1 UUUUGAUCCUUCUUUAGGUGCUCAAUUGGGCGCAGGUCAGGGUGCUUCUAACCAAAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCUACGGGUGCUCAAUUGGGCGCAGGUCAGGGUAU ((((((((((.....))((((((((((((((((.((((..(((.......)))...))))..(((((..((((........)))))))))..)))))))))))))))).))))))))... ( -50.80) >DroSim_CAF1 32238 120 + 1 AUUUGAUCCUUCUUUAGGUGCUCAAUUGGGCGCAGGUCAGGGUGUUUCCAACCAAAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCUGCGGGUGCUCAAUUGGGCGCAGGUCAGGGUAU .(((((((((.....))((((((((((((((((.((((..(((.......)))...))))..(..((..((((........))))))..)..)))))))))))))))).))))))).... ( -49.90) >DroYak_CAF1 33945 120 + 1 UUUUGAUCCCUCCUUGGGUGCUCAAUUGGGCGCAGGUCAGGGUGUUUCUAACCAAAGACCAAGUGGACAGGGAGCAUUUGAUCCUGCUACAGGUGCUCAAUUAGGCGCUGGUCAGGGUGU .......((((...(.(((((..((((((((((.((((..(((.......)))...))))..((((.(((((..(....).)))))))))..))))))))))..))))).)..))))... ( -49.10) >consensus UUUUGAUCCCUCCUUAGGUGCUCAAUUGGGCGCAGGUCAGGGUGUUUCUAACCAAAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCUACGGGUGCUCAAUUGGGCGCAGGUCAGGGUAU .......((((......((((((((((((((((.((((..(((.......)))...))))..(((((..((((........)))))))))..)))))))))))))))).....))))... (-33.34 = -36.70 + 3.36)

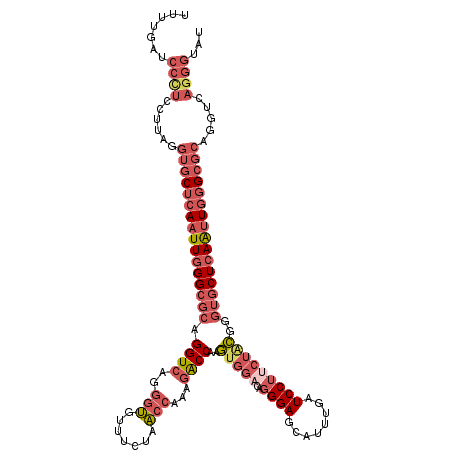
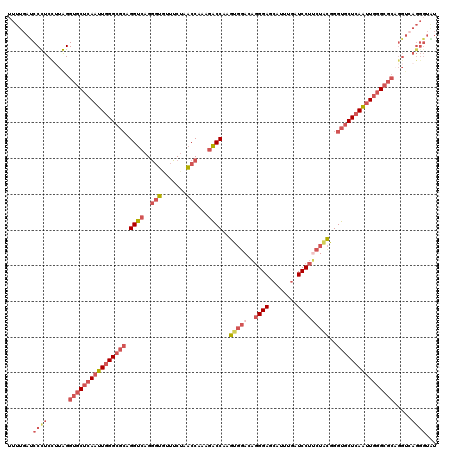
| Location | 4,455,137 – 4,455,254 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -43.59 |
| Consensus MFE | -21.08 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4455137 117 + 23771897 AUCCUUCUACAGGUGCUCAAUUGGGCGCAGGUGAGGGUGUUUCUAACCAAAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCUACGGGUGCUCAAUUGGGUGCAGGUCAGGGC---UUUA .((((...((..(..(((((((((((((.(((...(((.......)))....)))..(((((..((((........)))))))))..)))))))))))))..)..)).)))).---.... ( -44.80) >DroPse_CAF1 39741 111 + 1 AUCCCGCUCUGGGCGCUCAGCUGGGAGUUGGACAGGCUCCUCUUGGACAGAGGCCGAAUCAGC------CAAAUCCUCCUCCGUUCGGUGCUCAGCUUGGUGCUGGACAGGCGCCUC--- ..........(((((((......((((..(((..(((((((((.....))))).......)))------)...)))..))))((((((..((......))..)))))).))))))).--- ( -44.11) >DroSec_CAF1 32380 117 + 1 AUCCUUCUACGGGUGCUCAAUUGGGCGCAGGUCAGGGUAU---UAACCAGAGACCAAGUGCACAGGGAGCAUUUGAUCCUUCGACAGGUGCCCAACUAGGAGCUGGACAGGGUAUUUCUA (((((((((.....((((..((((((((..(((..(((..---..)))((.((.(((((((.......))))))).))))..)))..))))))))....)))))))).)))))....... ( -42.50) >DroSim_CAF1 32318 117 + 1 AUCCUUCUGCGGGUGCUCAAUUGGGCGCAGGUCAGGGUAU---UAACCAGAGACCAAGUGCACAGGGAGCAUUUGAUCCUUCGACAGGUGCCCAACUUGGAGCUGGACAGGGUAUUUCUA ....((((((.(((..((((((((((((..(((..(((..---..)))((.((.(((((((.......))))))).))))..)))..)))))))).)))).))).).)))))........ ( -41.90) >DroEre_CAF1 35173 118 + 1 --CCCUCCUUGGGUGCUCAAUUGGGAGCAGCUCAGGGUGUUCCUAACCAAAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCUACAGGUGCUCAACUUGGCGCUGGGCAGAGUGUUCCUA --..(..(((((...((...(((((((((.(....).)))))))))....)).)))))..)...((((((((((..(((..(..(((((.....)))))..)..)))..)))))))))). ( -43.20) >DroYak_CAF1 34025 120 + 1 AUCCUGCUACAGGUGCUCAAUUAGGCGCUGGUCAGGGUGUUUCUAACCAGAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCUACAGGUGCCCAACUUGGAACUGGACAGGGUGUUCCUA ...(((((((.((((((......))))))((((..(((.......)))...))))..)))).)))(((((((((..(((.....(((((.....))))).....)))..))))))))).. ( -45.00) >consensus AUCCUUCUACGGGUGCUCAAUUGGGCGCAGGUCAGGGUGUU_CUAACCAGAGACCAAGUGGACAGGGAGCAUUUGAUCCUUCGACAGGUGCCCAACUUGGAGCUGGACAGGGUAUUCCUA ..(((.....))).((((((((((((((.((((..(((.......)))...))))........(((((........)))))......)))))))).)))).))................. (-21.08 = -22.20 + 1.12)

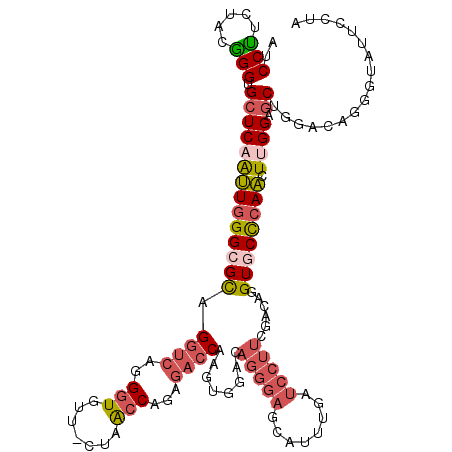
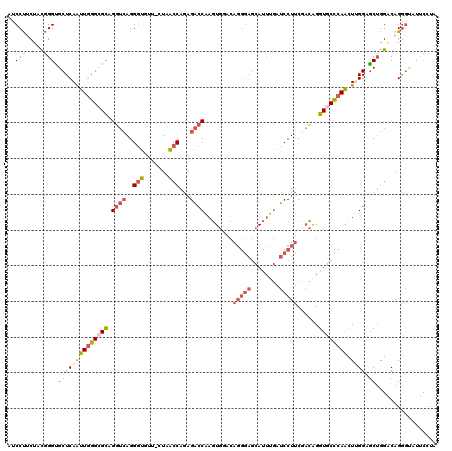
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:12 2006