| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,424,850 – 4,424,984 |
| Length | 134 |
| Max. P | 0.886888 |

| Location | 4,424,850 – 4,424,950 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.68 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
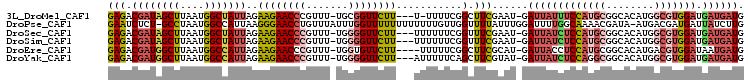
>3L_DroMel_CAF1 4424850 100 + 23771897 CAUCAUCAUCCACGCCAUGUGCCGCAUGGAAAUAAUC-AUUCGAAGCCGAAAA-A---AAGAACCGCA-AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUC ..............(((((.....)))))........-.((((....))))..-.---(((((((...-.....)))))))...(((((......)))))...... ( -19.80) >DroPse_CAF1 8375 104 + 1 CAAGAUAAUCAUCGUCAU-UAUCGUUUUGCCAAAAUCCAAAUAAAACCAACAAAAAAAAAAAACCAAAUAAACAGGUUCCCUUAAUGGCCAUUAGGC-AGAGAUUC ...((((((.......))-)))).(((((((..............................((((.........)))).((.....))......)))-)))).... ( -13.80) >DroSec_CAF1 6837 101 + 1 CAUCAUCAUCCACGCCAUGUGCCGCAUGGAGAUAAUC-AUUCGAAACCGAAAAAA---AAGAACCCCA-AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUC ..............(((((.....)))))((((....-.((((....))))....---((((((((..-....))))))))...(((((......))))).)))). ( -24.00) >DroSim_CAF1 4209 101 + 1 CAUCAUCAUCCACGCCAUGUGCCGCAUGGAGAUAAUC-AUUCGAAACCGAAAAAA---AAGAACCCCA-AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUC ..............(((((.....)))))((((....-.((((....))))....---((((((((..-....))))))))...(((((......))))).)))). ( -24.00) >DroEre_CAF1 6954 100 + 1 CAUCAUUAUCCACGUCAUGUGCCGCAUGGAGGUAAUC-AUGCGAAGCCGAAAAA----AAGAACACCA-AAACGGGUUCUUCUAAUGGCCAUUAAGCCAUCGUCUC ...........(((..(((.(((((((((......))-)))))..((((.....----((((((.((.-....))))))))....))))......))))))))... ( -24.10) >DroYak_CAF1 7909 101 + 1 CAUCAUCAUCCACGCCAUGUGCCGCCUGGAGAUAAUC-AUACGAAGCUGAAAAAU---AAGAACCCCA-AAACGGGUUCUUCUAAUGGCCAUUAAGCCAUCGUCUC ....(((.((((.((........)).)))))))....-..((((...........---((((((((..-....))))))))....((((......))))))))... ( -25.70) >consensus CAUCAUCAUCCACGCCAUGUGCCGCAUGGAGAUAAUC_AUUCGAAACCGAAAAAA___AAGAACCCCA_AAACGGGUUCUUCUAAUAGCCAUUAAGCUAUCGUCUC ........((((.((........)).))))...........(((..............((((((((.......))))))))....((((......))))))).... (-12.96 = -13.68 + 0.73)


| Location | 4,424,850 – 4,424,950 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -22.96 |
| Energy contribution | -24.22 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4424850 100 - 23771897 GAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUU-UGCGGUUCUU---U-UUUUCGGCUUCGAAU-GAUUAUUUCCAUGCGGCACAUGGCGUGGAUGAUGAUG ((((.(((((((....)))))))(((((((((.....-...)))))))---)-)))))..........-.(((((((((((((........))))))).)))))). ( -30.60) >DroPse_CAF1 8375 104 - 1 GAAUCUCU-GCCUAAUGGCCAUUAAGGGAACCUGUUUAUUUGGUUUUUUUUUUUUGUUGGUUUUAUUUGGAUUUUGGCAAAACGAUA-AUGACGAUGAUUAUCUUG .((((((.-(((....)))(((((((((((((.........))))))))..((((((..(..((.....))..)..))))))...))-)))..)).))))...... ( -20.20) >DroSec_CAF1 6837 101 - 1 GAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUU-UGGGGUUCUU---UUUUUUCGGUUUCGAAU-GAUUAUCUCCAUGCGGCACAUGGCGUGGAUGAUGAUG ((((((((((((....)))))))((((((((((....-..))))))))---))......)))))....-.(((((((((((((........))))))).)))))). ( -41.10) >DroSim_CAF1 4209 101 - 1 GAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUU-UGGGGUUCUU---UUUUUUCGGUUUCGAAU-GAUUAUCUCCAUGCGGCACAUGGCGUGGAUGAUGAUG ((((((((((((....)))))))((((((((((....-..))))))))---))......)))))....-.(((((((((((((........))))))).)))))). ( -41.10) >DroEre_CAF1 6954 100 - 1 GAGACGAUGGCUUAAUGGCCAUUAGAAGAACCCGUUU-UGGUGUUCUU----UUUUUCGGCUUCGCAU-GAUUACCUCCAUGCGGCACAUGACGUGGAUAAUGAUG ((((.(((((((....)))))))((((((((((....-.)).))))))----))))))....((((((-(........)))))))(((.....))).......... ( -29.50) >DroYak_CAF1 7909 101 - 1 GAGACGAUGGCUUAAUGGCCAUUAGAAGAACCCGUUU-UGGGGUUCUU---AUUUUUCAGCUUCGUAU-GAUUAUCUCCAGGCGGCACAUGGCGUGGAUGAUGAUG ((((.(((((((....)))))))..((((((((....-..))))))))---...))))..........-.((((((((((.((........)).)))).)))))). ( -36.50) >consensus GAGACGAUAGCUUAAUGGCCAUUAGAAGAACCCGUUU_UGGGGUUCUU___UUUUUUCGGCUUCGAAU_GAUUAUCUCCAUGCGGCACAUGGCGUGGAUGAUGAUG (((.((((((((....)))))))..((((((((.......))))))))...........).)))......(((((((((((((........))))))).)))))). (-22.96 = -24.22 + 1.26)
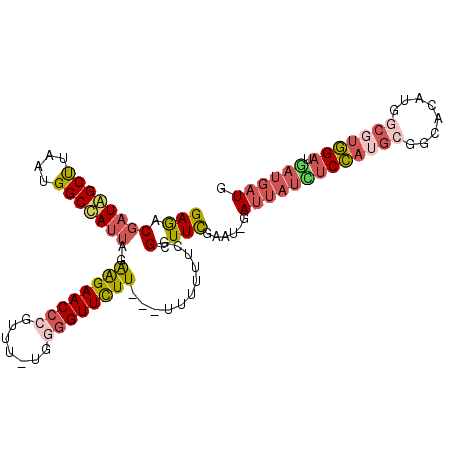

| Location | 4,424,889 – 4,424,984 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -16.91 |
| Energy contribution | -19.05 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4424889 95 - 23771897 GCAAAUGCUGAUUAUUAUGAUCGUAUUUGAGAGUGAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUUUGCGGUUCUUU-UUUUCGGCUUCGA .(((((((.((((.....))))))))))).((((((((.(((((((....)))))))(((((((((........))))))))-))))).))))... ( -27.20) >DroSec_CAF1 6876 96 - 1 GCAAAUUCUGAUUAUUAUGGUCGCAUUUGAGAGUGAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUUUGGGGUUCUUUUUUUUCGGUUUCGA .........((((.....))))....((((((.(((((.(((((((....)))))))((((((((((......)))))))))).))))).)))))) ( -29.30) >DroSim_CAF1 4248 96 - 1 GCAAAUGCUGAUUAUUAUGGUCGCAUUUGAGAGUGAGACGAUAGCUUAAUGGCUAUUAGAAGAACCCGUUUUGGGGUUCUUUUUUUUCGGUUUCGA .(((((((.((((.....)))))))))))....(((((((((((((....)))))))((((((((((......))))))))))......)))))). ( -35.10) >DroEre_CAF1 6993 95 - 1 GCAAAUGCUGAUUAUUAUGGUCGUAUUUGAGAGCGAGACGAUGGCUUAAUGGCCAUUAGAAGAACCCGUUUUGGUGUUCUU-UUUUUCGGCUUCGC .(((((((.((((.....)))))))))))...(((((.((((((((....)))))..((((((((((.....)).))))))-))..)))..))))) ( -30.70) >DroYak_CAF1 7948 96 - 1 GCAAAUGCUGAUUAUUAUAGUCGUAUUUGAGAGUGAGACGAUGGCUUAAUGGCCAUUAGAAGAACCCGUUUUGGGGUUCUUAUUUUUCAGCUUCGU .(((((((.(((((...)))))))))))).((((((((.(((((((....)))))))..((((((((......))))))))...))))).)))... ( -32.50) >DroAna_CAF1 7247 74 - 1 GCUAAUGCUGAUUAUUAUGGUCUUAGAA------GAGGCUUUAG-UUAAUGGCCAUUAGAAAAACCCGUUUGA------------UUUGGC---CG ((....))((((((....((((((....------)))))).)))-)))..((((((((((........)))))------------..))))---). ( -17.00) >consensus GCAAAUGCUGAUUAUUAUGGUCGUAUUUGAGAGUGAGACGAUAGCUUAAUGGCCAUUAGAAGAACCCGUUUUGGGGUUCUUUUUUUUCGGCUUCGA .(((((((.((((.....)))))))))))....(((((.(((((((....)))))))..((((((((......))))))))...)))))....... (-16.91 = -19.05 + 2.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:00 2006