
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,415,903 – 4,416,023 |
| Length | 120 |
| Max. P | 0.813180 |

| Location | 4,415,903 – 4,416,023 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -33.10 |
| Energy contribution | -32.02 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.72 |
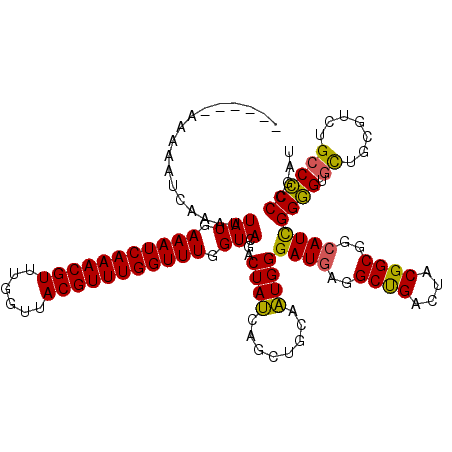
| Structure conservation index | 0.87 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
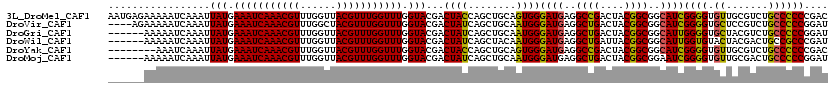
>3L_DroMel_CAF1 4415903 120 + 23771897 AAUGAGAAAAAUCAAAUUAUGAAAUCAAACGUUUGGUUACGUUUGGUUUGGUACGACUACCAGCUGCAGUGGGAUGAGGCCGACUACGGCGGCAUCGGGGUGUUGCGUCUGCCCCCCGAC ..(((......))).......(((((((((((......)))))))))))((((....)))).((((((((.((......)).)))...))))).((((((.((.......)).)))))). ( -40.20) >DroVir_CAF1 1 116 + 1 ----AGAAAAAUCAAAUUAUGAAAUCAAACGUUUGGCUACGUUUGGUUUGGUACGACUAUCAGCUGCAAUGGGAUGAGGCUGACUACGGCGGCAUCGGGGUGCUCCGUCUGCCCCCGGAU ----.............(((.(((((((((((......))))))))))).)))......((((((.((......)).))))))...(((.((((.((((....))))..)))).)))... ( -38.40) >DroGri_CAF1 1 114 + 1 ------AAAAAUCAAAUUAUGAAAUCAAACGUUUGGUUACGUUUGGUUUGGUACGACUAUCAGCUGCAAUGGGAUGAGGCUGACUACGGCGGCAUUGGGGUGCUACGUCUGCCCCCGGAU ------...........(((.(((((((((((......))))))))))).))).........(((((..(((...........)))..))))).((((((.((.......)))))))).. ( -35.80) >DroWil_CAF1 1 114 + 1 ------AAAAAUCAAAUUAUGAAAUCAAACGUUUGGUUACGUUUGGUUUGGUACGACUAUCAGCUACAAUGGGAUGAGGCUGAUUACGGCGGCAUUGGUGUACUACGACUGCCGCCCGAU ------...........(((.(((((((((((......))))))))))).)))((...(((((((.((......)).)))))))...((((((((((........))).))))))))).. ( -39.60) >DroYak_CAF1 1 112 + 1 --------AAAUCAAAUUAUGAAAUCAAACGUUUGGUUACGUUUGGUUUGGUACGACUACCAGCUGCAGUGGGAUGAGGCCGACUACGGCGGCAUCGGGGUGUUGCGUCUGCCCCCCGAC --------.............(((((((((((......)))))))))))((((....)))).((((((((.((......)).)))...))))).((((((.((.......)).)))))). ( -39.40) >DroMoj_CAF1 1 114 + 1 ------AAAAAUCAAAUUAUGAAAUCAAACGUUUGGUUACGUUUGGUUUGGUACGACUAUCAGCUGCAAUGGGAUGAGGCUGACUACGGCGGAAUCGGGGUGUUGCGACUGCCCCCGGAU ------.....((....(((.(((((((((((......))))))))))).)))((.((.((((((.((......)).))))))....)))))).((((((.((.......)))))))).. ( -34.70) >consensus ______AAAAAUCAAAUUAUGAAAUCAAACGUUUGGUUACGUUUGGUUUGGUACGACUAUCAGCUGCAAUGGGAUGAGGCUGACUACGGCGGCAUCGGGGUGCUGCGUCUGCCCCCCGAU .................(((.(((((((((((......))))))))))).)))...((((........))))((((..((((....))))..))))((((.((.......)))))).... (-33.10 = -32.02 + -1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:54 2006