| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,414,965 – 4,415,075 |
| Length | 110 |
| Max. P | 0.924529 |

| Location | 4,414,965 – 4,415,075 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
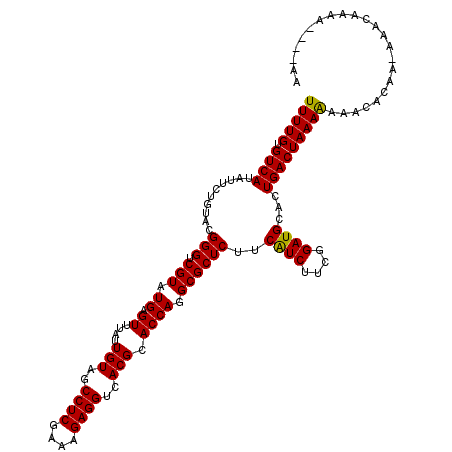
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -28.93 |
| Energy contribution | -28.57 |
| Covariance contribution | -0.36 |
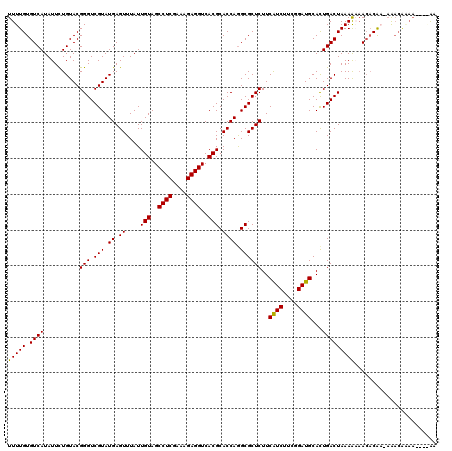
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4414965 110 - 23771897 UUUUGUGUCAUAUUCUGUACGGGUCGUAUGAGUUUAUUGUAGCCUCGAAAGAGGUCACGCACCAAGCGCUCUUCAUCUUCGGAUGCACUGACUAAAA-AACACAA-CG--------GAAA ............((((((..(((.(((.((.((....(((..((((....))))..))).)))).))))))..((((....))))............-......)-))--------))). ( -29.80) >DroVir_CAF1 17750 116 - 1 UUUUGUGUCAUAUUUUGUACGGGUCGUAUGAGUUUAUUGUAGCCUCGAAAGAGGUCACGCACCAGGCGCUCUUCGUCUUCGGAUGCACUGACUAAAGAAACACACGAAACGAAA----AA ((((((((......(..((((...))))..)((((...((..((((....))))..))((((((((((.....)))))..)).)))...........)))))))))))).....----.. ( -33.00) >DroPse_CAF1 14698 118 - 1 UUUUGUGUCAUAUUCUGUACGGGUCGUAUGAGUUUAUUGUAGCCUCGAAAGAGGUCACGCACCAGGCGCUCUUCAUCUUCGGAUGCACUGACUAAAAAAACACAA-AAACAAAAG-AGAA ((((((((........((.(((.......((((.....((..((((....))))..))((.....))))))..((((....))))..))))).......))))))-)).......-.... ( -31.96) >DroWil_CAF1 30335 119 - 1 UUUUGUGUCAUAUUCUGUACGGGUCGUAUGAGUUUAUUGUAGCCUCGAAAGAGGUCACGCACCAGGCGCUCUUCAUCUUCGGAUGCAUUGACUAAAAUUAACCGA-AAACGUAAUGGAAC ............(((((((((..(((...((((.....((..((((....))))..))((.....))))))..((((....)))).................)))-...))).)))))). ( -31.50) >DroMoj_CAF1 15259 114 - 1 UUUUGUGUCAUAUUUUGUACGGGUCGUAUGAGUUUAUUGUAGCCUCGAAAGAGGUCACGCACCAGGCGCUCUUCGUCUUCGGAUGCACUGACUAAAAAAACACA--AAACAGAA----GA ((((((((....((((.....((((....((((.....((..((((....))))..))((.....))))))..((((....))))....))))..)))))))))--))).....----.. ( -32.20) >DroPer_CAF1 15257 118 - 1 UUUUGUGUCAUAUUCUGUACGGGUCGUAUGAGUUUAUUGUAGCCUCGAAAGAGGUCACGCACCAGGCGCUCUUCAUCUUCGGAUGCACUGACUAAAAAAACACAA-AAACAAAAG-AGAA ((((((((........((.(((.......((((.....((..((((....))))..))((.....))))))..((((....))))..))))).......))))))-)).......-.... ( -31.96) >consensus UUUUGUGUCAUAUUCUGUACGGGUCGUAUGAGUUUAUUGUAGCCUCGAAAGAGGUCACGCACCAGGCGCUCUUCAUCUUCGGAUGCACUGACUAAAAAAACACAA_AAACAAAA____AA (((((.((((..........(((.(((.((.((....(((..((((....))))..))).)))).))))))..((((....))))...)))))))))....................... (-28.93 = -28.57 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:53 2006