| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,408,413 – 4,408,517 |
| Length | 104 |
| Max. P | 0.769305 |

| Location | 4,408,413 – 4,408,517 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641945 |
| Prediction | RNA |
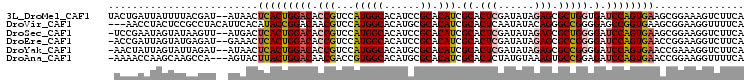
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4408413 104 + 23771897 UGAAGACUUUCCGCUUCACUGGAUCACCAGCGCUCUAUAUCGAGUGCGAUGUGCGGAUGUGCCAUGGACGGUGUCCAGUGAGUUAU--AUCGUAAAAUAAUCAGUA ..............(((((((((.((((.((((((......)))))).(((.(((....))))))....)))))))))))))....--.................. ( -33.30) >DroVir_CAF1 8819 103 + 1 UGAAAACCUUCCGCUUCACCGGCUCCCCGGCCCUGUAUAUUGAGUGCGAUGUGCGCAUGUGCCAUGGACGUUGUCCGGUAUGUGAAUGUAGGCGGAGUAGGUU--- ....((((((((((((((((((....))))....((((((((....)))))))).((((((((..((((...))))))))))))..)).)))))))..)))))--- ( -38.90) >DroSec_CAF1 6376 103 + 1 UGAAGACCUUCCGCUUCACUGGAUCCCCAGCGCUCUAUAUCGAGUGCGAUGUGCGGAUGUGCCAUGGACGGUGUCCAGUGAGUCAU--AACUUAUACUAUUCGGA- .........((((..((((((((..((..((((((......)))))).(((.(((....))))))....))..))))))))((...--.))..........))))- ( -31.80) >DroEre_CAF1 6603 103 + 1 UGAAGACCUUCCGGUUCACUGGAUCCCCGGCGCUCUAUAUCGAGUGCGAUGUGCGGAUGUGCCAUGGACGGUGUCCAGUGAGUUUC--AUCUCAUACUAAUCGGU- ((((((((....)))(((((((((((((((((((((((((((....))))))..))).)))))..))..)).))))))))).))))--)................- ( -34.10) >DroYak_CAF1 6675 103 + 1 UGAAGACCUUUCGGUUCACUGGAUCCCCGGCGCUCUAUAUCGAGUGCGAUGUGCGCAUGUGCCAUGGACGGUGUCCAGUGAGUUAU--AUCUAAUACUAAUAGUU- .....(((....)))(((((((((..(((((((((......)))))).(((.(((....))))))...))).))))))))).....--.................- ( -33.20) >DroAna_CAF1 7014 102 + 1 UGAAAACCUUCCGGUUCACUGGAUCUCCGGCACUUUACAUAGAGUGCGAUGUGCGCAUGUGCCACGGUCGUUGUCCAGUAAGUACU---UGGCUUGCUUGGUUUU- ..((((((.(((((....)))))...((.(((((((....)))))))...(((.((....))))))).........(((((((...---..))))))).))))))- ( -30.90) >consensus UGAAGACCUUCCGCUUCACUGGAUCCCCGGCGCUCUAUAUCGAGUGCGAUGUGCGCAUGUGCCAUGGACGGUGUCCAGUGAGUUAU__AUCGCAUACUAAUCGGU_ ..............((((((((((..(((((((((......))))))..((.(((....)))))))).....))))))))))........................ (-23.86 = -24.00 + 0.14)

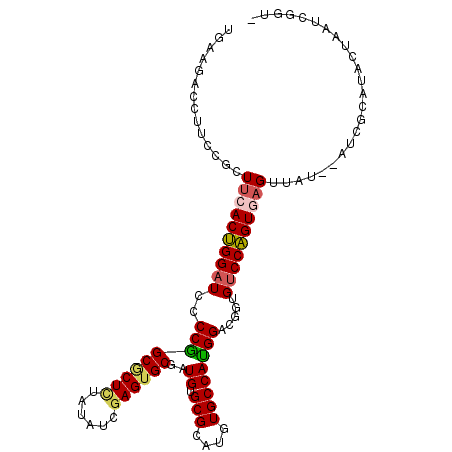

| Location | 4,408,413 – 4,408,517 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4408413 104 - 23771897 UACUGAUUAUUUUACGAU--AUAACUCACUGGACACCGUCCAUGGCACAUCCGCACAUCGCACUCGAUAUAGAGCGCUGGUGAUCCAGUGAAGCGGAAAGUCUUCA ....((..(((((.((..--.....((((((((((((....(((((......)).))).((.(((......))).)).)))).))))))))..)).)))))..)). ( -31.70) >DroVir_CAF1 8819 103 - 1 ---AACCUACUCCGCCUACAUUCACAUACCGGACAACGUCCAUGGCACAUGCGCACAUCGCACUCAAUAUACAGGGCCGGGGAGCCGGUGAAGCGGAAGGUUUUCA ---(((((..(((((.............((((((...))))........((((.....))))...........))(((((....)))))...)))))))))).... ( -31.30) >DroSec_CAF1 6376 103 - 1 -UCCGAAUAGUAUAAGUU--AUGACUCACUGGACACCGUCCAUGGCACAUCCGCACAUCGCACUCGAUAUAGAGCGCUGGGGAUCCAGUGAAGCGGAAGGUCUUCA -((((....(((.....)--))...((((((((..((....(((((......)).))).((.(((......))).))..))..))))))))..))))......... ( -30.00) >DroEre_CAF1 6603 103 - 1 -ACCGAUUAGUAUGAGAU--GAAACUCACUGGACACCGUCCAUGGCACAUCCGCACAUCGCACUCGAUAUAGAGCGCCGGGGAUCCAGUGAACCGGAAGGUCUUCA -................(--(((..((((((((..((....(((((......)).))).((.(((......))).))..))..))))))))(((....))).)))) ( -31.80) >DroYak_CAF1 6675 103 - 1 -AACUAUUAGUAUUAGAU--AUAACUCACUGGACACCGUCCAUGGCACAUGCGCACAUCGCACUCGAUAUAGAGCGCCGGGGAUCCAGUGAACCGAAAGGUCUUCA -..(((.......)))..--.....((((((((.....(((.((((...((((.....))))(((......))).)))).)))))))))))(((....)))..... ( -32.60) >DroAna_CAF1 7014 102 - 1 -AAAACCAAGCAAGCCA---AGUACUUACUGGACAACGACCGUGGCACAUGCGCACAUCGCACUCUAUGUAAAGUGCCGGAGAUCCAGUGAACCGGAAGGUUUUCA -................---.....(((((((((..((...(((((....)).)))...(((((........)))))))..).))))))))(((....)))..... ( -28.60) >consensus _ACCGAUUAGUAUGAGAU__AUAACUCACUGGACACCGUCCAUGGCACAUCCGCACAUCGCACUCGAUAUAGAGCGCCGGGGAUCCAGUGAACCGGAAGGUCUUCA .........................(((((((((.(((...(((((......)).))).((.(((......))).))))).).))))))))............... (-20.11 = -20.25 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:49 2006