
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,406,678 – 4,406,768 |
| Length | 90 |
| Max. P | 0.857277 |

| Location | 4,406,678 – 4,406,768 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -14.08 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
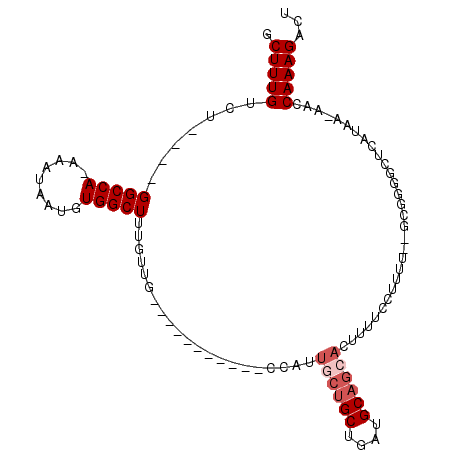
>3L_DroMel_CAF1 4406678 90 + 23771897 GCUUUGUCU----GGCCA-AAAUAAUGUGGCUUUGUUG-----------CCAUUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCGAGGCUCAUAA-AGCCAAAGACU (((((((..----((((.-.......(((((......)-----------))))((((((....)))))).................)))).))))-)))........ ( -29.20) >DroPse_CAF1 5459 105 + 1 GCUUUGUCUGCCUGGCCAAAAAUUAUGUGGCUUUGUGGCUGUUGCCUAGCCUCUUCUGCUGAUGCAGCACUUUUCGUUGU--GCGGGGCUCAUAAAAACCAAAGACU .....((((((..(((((.........)))))..))(((....))).((((((...(((....)))((((........))--))))))))............)))). ( -31.60) >DroEre_CAF1 4871 90 + 1 GCUUUGUCU----GGCCA-AAAUAAUGUGGCUUUGUUG-----------CCAUUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCAGGGCUCAUAA-AGCCAAAGACU (((((((..----((((.-.......(((((......)-----------))))((((((....)))))).................)))).))))-)))........ ( -29.00) >DroYak_CAF1 4938 95 + 1 GCUUUGUCU----GGCCA-AAAUAAUGUGGCUUUGUUGAUGUG------GCAUUGCUGCUGAUGCAGCACUUUUCCUUUUUCGCGGCGCUCAUAA-AACCAAAGACU .....((((----(((((-........)))))..(((..((((------((..((((((....)))))).....((........)).)).)))).-)))...)))). ( -24.70) >DroAna_CAF1 5314 70 + 1 GCUUUGUCU---UGGCCA-AAAUUAUGUGGCUUUGUU--------------------GCUGAUGCAGCACUUUUGUU------------UCAUAA-AACCAAAGACU .....((((---((((((-........))))).((((--------------------((....))))))........------------......-.....))))). ( -17.40) >DroPer_CAF1 5745 105 + 1 GCUUUGUCUGCCUGGCCAAAAAUUAUGUGGCUUUGUGGCUGGUGCCUAGCCUCUGCUGCUGAUGCAGCACUUUUCGUUGU--GCGGGGCUCAUAAAAACCAAAGACU ..(((((..((((.(((((.......(.((((....(((....))).)))).)((((((....)))))).......))).--)).))))..)))))........... ( -35.40) >consensus GCUUUGUCU____GGCCA_AAAUAAUGUGGCUUUGUUG___________CCAUUGCUGCUGAUGCAGCACUUUUCCUUUU__GCGGGGCUCAUAA_AACCAAAGACU .(((((.......(((((.........))))).....................((((((....))))))..............................)))))... (-14.08 = -14.92 + 0.83)

| Location | 4,406,678 – 4,406,768 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -13.27 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
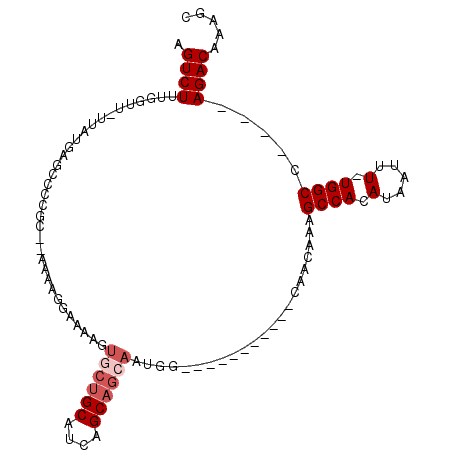
>3L_DroMel_CAF1 4406678 90 - 23771897 AGUCUUUGGCU-UUAUGAGCCUCGCGAAAAAGGAAAAGUGCUGCAUCAGCAGCAAUGG-----------CAACAAAGCCACAUUAUUU-UGGCC----AGACAAAGC .((((..((((-(...)))))..((.((((........((((((....)))))).(((-----------(......)))).....)))-).)).----))))..... ( -27.90) >DroPse_CAF1 5459 105 - 1 AGUCUUUGGUUUUUAUGAGCCCCGC--ACAACGAAAAGUGCUGCAUCAGCAGAAGAGGCUAGGCAACAGCCACAAAGCCACAUAAUUUUUGGCCAGGCAGACAAAGC .((((((((((.(((((((((.(((--((........))))(((....)))...).)))).(((....))).........))))).....))))))..))))..... ( -30.00) >DroEre_CAF1 4871 90 - 1 AGUCUUUGGCU-UUAUGAGCCCUGCGAAAAAGGAAAAGUGCUGCAUCAGCAGCAAUGG-----------CAACAAAGCCACAUUAUUU-UGGCC----AGACAAAGC .((((..((((-..((((..(((.......))).....((((((....)))))).(((-----------(......))))..))))..-.))))----))))..... ( -27.80) >DroYak_CAF1 4938 95 - 1 AGUCUUUGGUU-UUAUGAGCGCCGCGAAAAAGGAAAAGUGCUGCAUCAGCAGCAAUGC------CACAUCAACAAAGCCACAUUAUUU-UGGCC----AGACAAAGC .((((.(((((-((.(((((...))......((.....((((((....))))))...)------)...)))..)))))))((......-))...----))))..... ( -22.80) >DroAna_CAF1 5314 70 - 1 AGUCUUUGGUU-UUAUGA------------AACAAAAGUGCUGCAUCAGC--------------------AACAAAGCCACAUAAUUU-UGGCCA---AGACAAAGC .(((((.((((-(((((.------------........(((((...))))--------------------).........)))))...-.)))))---))))..... ( -16.17) >DroPer_CAF1 5745 105 - 1 AGUCUUUGGUUUUUAUGAGCCCCGC--ACAACGAAAAGUGCUGCAUCAGCAGCAGAGGCUAGGCACCAGCCACAAAGCCACAUAAUUUUUGGCCAGGCAGACAAAGC .((((..(((((....)))))..((--....((((((.((((((....))))))..((((.(((....)))....))))......)))))).....))))))..... ( -32.50) >consensus AGUCUUUGGUU_UUAUGAGCCCCGC__AAAAGGAAAAGUGCUGCAUCAGCAGCAAUGG___________CAACAAAGCCACAUAAUUU_UGGCC____AGACAAAGC .((((.................................((((((....))))))......................((((.(.....).)))).....))))..... (-13.27 = -14.10 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:48 2006