
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,401,751 – 4,401,895 |
| Length | 144 |
| Max. P | 0.999121 |

| Location | 4,401,751 – 4,401,871 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -36.18 |
| Energy contribution | -36.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4401751 120 - 23771897 GAUCACCAACCUUGGAUGCUCAAUCAAUAGAUUCUCAAAUGGUGAUGUUUCCAUAUUGAUUGGCCCAAUCGAGGUGAAGCAUGGUGGCGGUGUUACGUACCAGGACUCCUCCAUCAGCAC ..(((((....((((..((.(((((((((((..((((.....))).)..))..)))))))))))))))....))))).((.((((((.((.(((.(......)))).)).)))))))).. ( -38.10) >DroVir_CAF1 82136 120 - 1 GAUCACCAACCUUGGAUGCUCAAUCAAUAGAUUCUCAAAGGGUGAUGCUUCCAUAUUGAUUGGCCCAAUCGAGGUGAAGCAUGGUGGCGGUGUUACGUACCAGGACUCCUCCAUCAGCAC ..(((((....((((..((.((((((((((.....)...(((.......))).)))))))))))))))....))))).((.((((((.((.(((.(......)))).)).)))))))).. ( -37.80) >DroPse_CAF1 57859 120 - 1 GAUCACCAACCUUGGAUGCUCAAUCAAUAGAUUCUCAAAUGGUGAAGUUUCCAUAUUGAUUGGCCCAAUCGAGGUGAAGCAUGGUGGCGGUGUUACGUACCAGGACUCCUCCAUCAGCAC ..(((((....((((..((.(((((((((((..((((.....)).))..))..)))))))))))))))....))))).((.((((((.((.(((.(......)))).)).)))))))).. ( -38.10) >DroGri_CAF1 67835 120 - 1 AAUCACCAACCUUGGAUGCUCAAUCAAUAGAUUCUCAAAGGGUGAUGUUUCCAUAUUGAUUGGCCCAAUCGAGGUGAAGCAUGGUGGCGGUGUUACGUACCAGGACUCCUCCAUCAUAAC ..(((((....((((..((.(((((((((((..((((.....))).)..))..)))))))))))))))....)))))...(((((((.((.(((.(......)))).)).)))))))... ( -36.10) >DroMoj_CAF1 71941 120 - 1 AAUCACCAACCUUGGAUGCUCAAUCAAUAGAUUCUCAAAGGGUGAUGCUUCCAUAUUGAUUGGCCCAAUCGAGGUGAAGCAUGGUGGCGGUGUUACGUACCAGGACUCCUCCAUCAGCAG ..(((((....((((..((.((((((((((.....)...(((.......))).)))))))))))))))....))))).((.((((((.((.(((.(......)))).)).)))))))).. ( -37.80) >DroPer_CAF1 57774 120 - 1 GAUCACCAACCUUGGAUGCUCAAUCAAUAGAUUCUCAAAUGGUGAAGUUUCCAUAUUGAUUGGCCCAAUCGAGGUGAAGCAUGGUGGCGGUGUUACGUACCAGGACUCCUCCAUCAGCAC ..(((((....((((..((.(((((((((((..((((.....)).))..))..)))))))))))))))....))))).((.((((((.((.(((.(......)))).)).)))))))).. ( -38.10) >consensus GAUCACCAACCUUGGAUGCUCAAUCAAUAGAUUCUCAAAGGGUGAUGUUUCCAUAUUGAUUGGCCCAAUCGAGGUGAAGCAUGGUGGCGGUGUUACGUACCAGGACUCCUCCAUCAGCAC ..(((((....((((..((.(((((((((((..((((.....))).)..))..)))))))))))))))....))))).((.((((((.((.(((.(......)))).)).)))))))).. (-36.18 = -36.52 + 0.33)


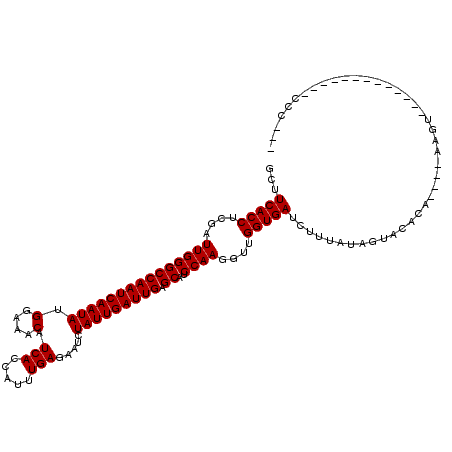
| Location | 4,401,791 – 4,401,895 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -23.17 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4401791 104 + 23771897 GCUUCACCUCGAUUGGGCCAAUCAAUAUGGAAACAUCACCAUUUGAGAAUCUAUUGAUUGAGCAUCCAAGGUUGGUGAUCUUUAUAGUACACAUUACAAGU-------------CUC--- ...(((((....(((((((((((((((((....))(((.....))).....))))))))).))..))))....))))).......................-------------...--- ( -25.00) >DroVir_CAF1 82176 101 + 1 GCUUCACCUCGAUUGGGCCAAUCAAUAUGGAAGCAUCACCCUUUGAGAAUCUAUUGAUUGAGCAUCCAAGGUUGGUGAUCUUUGUUGUACCCC----ACC--------------CCAAA- ............(((((.....(((((.(((..(((((.((((.((...((........))...)).)))).))))).))).)))))......----..)--------------)))).- ( -23.62) >DroPse_CAF1 57899 102 + 1 GCUUCACCUCGAUUGGGCCAAUCAAUAUGGAAACUUCACCAUUUGAGAAUCUAUUGAUUGAGCAUCCAAGGUUGGUGAUCUUUAUAGUACACAUUU---GU-------------CCC--U ...(((((....(((((((((((((((.(....)((((.....))))....))))))))).))..))))....)))))..................---..-------------...--. ( -24.10) >DroGri_CAF1 67875 104 + 1 GCUUCACCUCGAUUGGGCCAAUCAAUAUGGAAACAUCACCCUUUGAGAAUCUAUUGAUUGAGCAUCCAAGGUUGGUGAUUUUUGUUGUACUUA---CAACU-------------CUUCAA ...(((((....(((((((((((((((((....))(((.....))).....))))))))).))..))))....))))).....(((((....)---)))).-------------...... ( -28.50) >DroMoj_CAF1 71981 114 + 1 GCUUCACCUCGAUUGGGCCAAUCAAUAUGGAAGCAUCACCCUUUGAGAAUCUAUUGAUUGAGCAUCCAAGGUUGGUGAUUUUC--UCUACCUC---CAAAACAAAAACAAAAAACCAAA- ...(((((....((((((((((((..(((((..(.(((.....))))..))))))))))).))..))))....))))).....--........---.......................- ( -23.60) >DroPer_CAF1 57814 102 + 1 GCUUCACCUCGAUUGGGCCAAUCAAUAUGGAAACUUCACCAUUUGAGAAUCUAUUGAUUGAGCAUCCAAGGUUGGUGAUCUUUAUAGUACACAUUU---GU-------------CCC--U ...(((((....(((((((((((((((.(....)((((.....))))....))))))))).))..))))....)))))..................---..-------------...--. ( -24.10) >consensus GCUUCACCUCGAUUGGGCCAAUCAAUAUGGAAACAUCACCAUUUGAGAAUCUAUUGAUUGAGCAUCCAAGGUUGGUGAUCUUUAUAGUACACA____AAGU_____________CCC___ ...(((((....(((((((((((((((.(....).(((.....))).....))))))))).))..))))....))))).......................................... (-23.17 = -23.17 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:45 2006