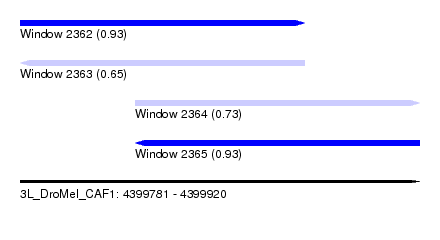
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,399,781 – 4,399,920 |
| Length | 139 |
| Max. P | 0.927656 |

| Location | 4,399,781 – 4,399,880 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -15.01 |
| Energy contribution | -15.81 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4399781 99 + 23771897 UUCACUUUCGCAAUCGAAUUCGAAAGCCCAUUUGCAUUGAUUUCGCUUGAGCUAUAUAU---UGUAUUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAU .........((((.......(....).....))))((((...(((((((.(((((((..---..........)))))))...)))))))..))))....... ( -20.50) >DroSec_CAF1 49708 94 + 1 UUCACUUUCGCAAUCGAAUUCGAAAGCCCAUUUGCAUUGAUUUCGCUUGAGCUAUAUA--------UUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAU .........((((.......(....).....))))((((...(((((((.(((((((.--------......)))))))...)))))))..))))....... ( -21.30) >DroSim_CAF1 50923 94 + 1 UUCACUUUUGCAAUCGAAUUCGAAAGCCCAUUUGCAUUGAUUUCGCUUGAGCUAUAUA--------UUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAU .........((((.......(....).....))))((((...(((((((.(((((((.--------......)))))))...)))))))..))))....... ( -21.00) >DroEre_CAF1 51832 94 + 1 UUCACUUUCGCAAUCGAAUUUGAAUGGUCAUUUGCAUUGAUUUCGCUGGAGCUAUAU--------AUUUAUAAUAUAGCAUGGAAGUGAUGCAAUAAAAAAU .(((((((((((..((((..(.((((........)))).).)))).))..(((((((--------.......))))))).)))))))))............. ( -20.60) >DroYak_CAF1 52899 89 + 1 UUCACUUUCGCAAUCGAAU-------------UGCAUUGAUUUCGCUUGAGCUAUAUACAUACAUACUUAUAAUAAAGCAUCGAAGUGAUGCAAUAAAAAAU .........(((((...))-------------)))((((...(((((((((((.(((...............))).))).)).))))))..))))....... ( -14.76) >consensus UUCACUUUCGCAAUCGAAUUCGAAAGCCCAUUUGCAUUGAUUUCGCUUGAGCUAUAUA_______AUUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAU .........((((..................))))((((...(((((((((((((((...............))))))).)).))))))..))))....... (-15.01 = -15.81 + 0.80)



| Location | 4,399,781 – 4,399,880 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -19.58 |
| Consensus MFE | -13.70 |
| Energy contribution | -14.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4399781 99 - 23771897 AUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAAUACA---AUAUAUAGCUCAAGCGAAAUCAAUGCAAAUGGGCUUUCGAAUUCGAUUGCGAAAGUGAA ((((((..(((((((.(((((..(((((((..........---..)))))))))))).))......)))))....((..(((....)))..))))))))... ( -21.10) >DroSec_CAF1 49708 94 - 1 AUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAA--------UAUAUAGCUCAAGCGAAAUCAAUGCAAAUGGGCUUUCGAAUUCGAUUGCGAAAGUGAA ((((((..(((((((.(((((..(((((((......--------.)))))))))))).))......)))))....((..(((....)))..))))))))... ( -21.90) >DroSim_CAF1 50923 94 - 1 AUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAA--------UAUAUAGCUCAAGCGAAAUCAAUGCAAAUGGGCUUUCGAAUUCGAUUGCAAAAGUGAA ((((((..(((((((.(((((..(((((((......--------.)))))))))))).))......)))))....((..(((....)))..))))))))... ( -21.50) >DroEre_CAF1 51832 94 - 1 AUUUUUUAUUGCAUCACUUCCAUGCUAUAUUAUAAAU--------AUAUAGCUCCAGCGAAAUCAAUGCAAAUGACCAUUCAAAUUCGAUUGCGAAAGUGAA .............((((((.(..(((((((.......--------)))))))...............((((.(((..........))).))))).)))))). ( -16.90) >DroYak_CAF1 52899 89 - 1 AUUUUUUAUUGCAUCACUUCGAUGCUUUAUUAUAAGUAUGUAUGUAUAUAGCUCAAGCGAAAUCAAUGCA-------------AUUCGAUUGCGAAAGUGAA .......(((((((...((((..(((.(((((((......)))).))).))).....))))....)))))-------------))....(..(....)..). ( -16.50) >consensus AUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAAU_______UAUAUAGCUCAAGCGAAAUCAAUGCAAAUGGGCUUUCGAAUUCGAUUGCGAAAGUGAA ........(((((((.((((...(((((((...............))))))).)))).))......)))))..................(..(....)..). (-13.70 = -14.70 + 1.00)



| Location | 4,399,821 – 4,399,920 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -20.69 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4399821 99 + 23771897 UUUCGCUUGAGCUAUAUAU---UGUAUUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAUAAUUGCUAUCAUUUUGUGGGGAACUUGUCGUUUGCGUUUU ...(((.((((((((((..---..........))))))).(((((((((((((((........))))).))))))...)))).......)))...))).... ( -21.80) >DroSec_CAF1 49748 94 + 1 UUUCGCUUGAGCUAUAUA--------UUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAUAAUUGCCAUCCUUUUGUAGGGAACUUGUCGUUUGCGUUUU ...(((.((((((((((.--------......)))))))...(((((...(((((........)))))..(((((....)))))))))))))...))).... ( -22.50) >DroSim_CAF1 50963 94 + 1 UUUCGCUUGAGCUAUAUA--------UUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAUAAUUGCCAUCCUUUUGUAGGGAACUUGUCGUUUGCGUUUU ...(((.((((((((((.--------......)))))))...(((((...(((((........)))))..(((((....)))))))))))))...))).... ( -22.50) >DroEre_CAF1 51872 94 + 1 UUUCGCUGGAGCUAUAU--------AUUUAUAAUAUAGCAUGGAAGUGAUGCAAUAAAAAAUAAUUGCCAUCCUUGUGUGGGCAACUUCUCUUUUGUGUUUU ...(((.((((((((((--------.......)))))))...(((((...(((((........)))))..(((......)))..))))))))...))).... ( -19.30) >DroYak_CAF1 52926 102 + 1 UUUCGCUUGAGCUAUAUACAUACAUACUUAUAAUAAAGCAUCGAAGUGAUGCAAUAAAAAAUAAUUGCCAUCCUUAUGUAGAGAACUGCUCUUUUGUGUUUU ...(((..((((........(((((.(((......))).....(((.((((((((........)))).)))))))))))).......))))....))).... ( -17.36) >consensus UUUCGCUUGAGCUAUAUA_______AUUUAUAAUAUAGCAUCCAAGUGAUGCAAUAAAAAAUAAUUGCCAUCCUUUUGUAGGGAACUUGUCGUUUGCGUUUU ...(((.((((((((((...............)))))))...(((((...(((((........)))))..(((((....)))))))))))))...))).... (-16.20 = -16.20 + -0.00)


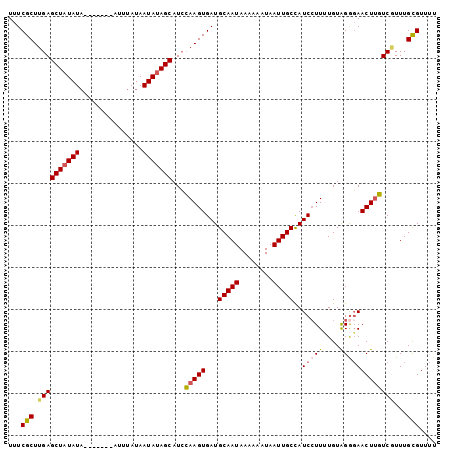
| Location | 4,399,821 – 4,399,920 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -14.24 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4399821 99 - 23771897 AAAACGCAAACGACAAGUUCCCCACAAAAUGAUAGCAAUUAUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAAUACA---AUAUAUAGCUCAAGCGAAA ....(((......(((((............(((.(((((........)))))))))))))((.(((((((..........---..)))))))))..)))... ( -19.50) >DroSec_CAF1 49748 94 - 1 AAAACGCAAACGACAAGUUCCCUACAAAAGGAUGGCAAUUAUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAA--------UAUAUAGCUCAAGCGAAA ....(((......(((((..(((.....)))...(((((........)))))...)))))((.(((((((......--------.)))))))))..)))... ( -22.80) >DroSim_CAF1 50963 94 - 1 AAAACGCAAACGACAAGUUCCCUACAAAAGGAUGGCAAUUAUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAA--------UAUAUAGCUCAAGCGAAA ....(((......(((((..(((.....)))...(((((........)))))...)))))((.(((((((......--------.)))))))))..)))... ( -22.80) >DroEre_CAF1 51872 94 - 1 AAAACACAAAAGAGAAGUUGCCCACACAAGGAUGGCAAUUAUUUUUUAUUGCAUCACUUCCAUGCUAUAUUAUAAAU--------AUAUAGCUCCAGCGAAA ........((((((.(((((((..(....)...))))))).))))))......((.((.....(((((((.......--------)))))))...)).)).. ( -16.90) >DroYak_CAF1 52926 102 - 1 AAAACACAAAAGAGCAGUUCUCUACAUAAGGAUGGCAAUUAUUUUUUAUUGCAUCACUUCGAUGCUUUAUUAUAAGUAUGUAUGUAUAUAGCUCAAGCGAAA ...........((((.((....((((((..((((.((((........))))))))......((((((......)))))).)))))).)).))))........ ( -19.80) >consensus AAAACGCAAACGACAAGUUCCCUACAAAAGGAUGGCAAUUAUUUUUUAUUGCAUCACUUGGAUGCUAUAUUAUAAAU_______UAUAUAGCUCAAGCGAAA ....(((......(((((............(((.(((((........)))))))))))))((.(((((((...............)))))))))..)))... (-14.24 = -14.76 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:43 2006