
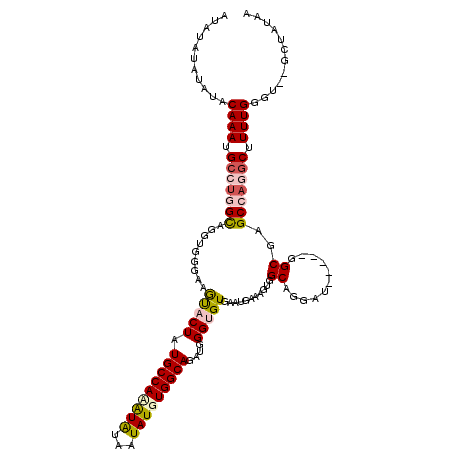
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,356,683 – 4,356,801 |
| Length | 118 |
| Max. P | 0.974521 |

| Location | 4,356,683 – 4,356,801 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -24.68 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4356683 118 + 23771897 UUAUAUAUAUACAAAUGCCUGGCAGGUGGGAAGCACUAUGCCAUAUAUAAUAUGUGGCAGAUGGGUGUGAAUGGAUGUGGCAGGAGCAAGGAGCGAGCCAGGCUUUUGGGU--GCUAUAA ...((((((..((((.(((((((.........(((((.(((((((((...)))))))))....))))).......(((.((....)).....))).))))))).))))..)--).)))). ( -38.80) >DroSec_CAF1 7354 115 + 1 AUAUAUAUAUACAAAUGCCUGGUAGGUGGGAAGUUCUAUGCCACAUAUAAUAUGUGGCAGAUGGGUGUGAAUGAAAGUGGCAGGAU-----GGCGAGCCAGGCUUUUGGGUUGGCUAUAA .((((..((..((((.(((((((..((.(...(..(..(((((((((...)))))))))...)..)((..((....)).))....)-----.))..))))))).))))...))..)))). ( -32.80) >DroSim_CAF1 7325 115 + 1 AUAUAUAUAUACAAAUGCCUGGCAGGUGGCAAGUUCUAUGCCACGUAUAAUAUGUGGCAGAUGGGUGUGAAUGAAAGUGGCAGGAU-----GGCGAGCCAGGCUUUUGGGUUGGCUAUAA .((((..((..((((.(((((((...((.((.(..(..(((((((((...)))))))))...)..)((..((....)).))....)-----).)).))))))).))))...))..)))). ( -35.50) >DroEre_CAF1 7389 107 + 1 AUA------UACAAAUGGCAGGCAGGUGGGAAACACUAUGCCAAAUGUAAUAUAUGGCAGAUGGGUGUGAAUAACAGUGGCAGGAU-----GGCAAGCAAAGCUUUUGGGU--GCUAUAA ...------((((..(((((....((((.....)))).)))))..))))...((((((.....(.(((.....))).).((..((.-----(((.......))).))..))--)))))). ( -25.00) >DroYak_CAF1 7416 113 + 1 AUAUAUAAAUACAAAUGCCUGGCAGGUGGGAAGUACUAUGCCAAAUGUAAUAUAUGGCAGAUGGGUGUGAAUAACGGUGGCAGGAU-----GGCAGGCAAAGCUUUUGGGU--GCUAUAA ...............((((((.((.((((......))))((((..(((.((.((((.(.....).)))).)).))).))))....)-----).)))))).(((........--))).... ( -25.80) >consensus AUAUAUAUAUACAAAUGCCUGGCAGGUGGGAAGUACUAUGCCAAAUAUAAUAUGUGGCAGAUGGGUGUGAAUGAAAGUGGCAGGAU_____GGCGAGCCAGGCUUUUGGGU__GCUAUAA ...........((((.(((((((.........(((((.(((((((((...)))))))))....)))))...........((...........))..))))))).))))............ (-24.68 = -25.64 + 0.96)


| Location | 4,356,683 – 4,356,801 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -15.96 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4356683 118 - 23771897 UUAUAGC--ACCCAAAAGCCUGGCUCGCUCCUUGCUCCUGCCACAUCCAUUCACACCCAUCUGCCACAUAUUAUAUAUGGCAUAGUGCUUCCCACCUGCCAGGCAUUUGUAUAUAUAUAA .((((..--...((((.(((((((..((.....))..................(((.....(((((.(((...))).)))))..)))..........))))))).))))....))))... ( -26.00) >DroSec_CAF1 7354 115 - 1 UUAUAGCCAACCCAAAAGCCUGGCUCGCC-----AUCCUGCCACUUUCAUUCACACCCAUCUGCCACAUAUUAUAUGUGGCAUAGAACUUCCCACCUACCAGGCAUUUGUAUAUAUAUAU .((((.......((((.((((((...((.-----.....))....................(((((((((...)))))))))................)))))).))))......)))). ( -25.92) >DroSim_CAF1 7325 115 - 1 UUAUAGCCAACCCAAAAGCCUGGCUCGCC-----AUCCUGCCACUUUCAUUCACACCCAUCUGCCACAUAUUAUACGUGGCAUAGAACUUGCCACCUGCCAGGCAUUUGUAUAUAUAUAU .((((.......((((.(((((((..((.-----.....))...................................((((((.......))))))..))))))).))))......)))). ( -27.42) >DroEre_CAF1 7389 107 - 1 UUAUAGC--ACCCAAAAGCUUUGCUUGCC-----AUCCUGCCACUGUUAUUCACACCCAUCUGCCAUAUAUUACAUUUGGCAUAGUGUUUCCCACCUGCCUGCCAUUUGUA------UAU ..(((((--(.....((((...))))((.-----.....))...)))))).....................((((..((((((((.((.....)))))..)))))..))))------... ( -15.40) >DroYak_CAF1 7416 113 - 1 UUAUAGC--ACCCAAAAGCUUUGCCUGCC-----AUCCUGCCACCGUUAUUCACACCCAUCUGCCAUAUAUUACAUUUGGCAUAGUACUUCCCACCUGCCAGGCAUUUGUAUUUAUAUAU ....(((--........))).((((((.(-----(..........((.....)).......(((((...........)))))..............)).))))))............... ( -16.10) >consensus UUAUAGC__ACCCAAAAGCCUGGCUCGCC_____AUCCUGCCACUUUCAUUCACACCCAUCUGCCACAUAUUAUAUGUGGCAUAGUACUUCCCACCUGCCAGGCAUUUGUAUAUAUAUAU ............((((.(((((((..((...........))....................(((((...........)))))...............))))))).))))........... (-15.96 = -16.72 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:37 2006