
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,353,754 – 4,353,844 |
| Length | 90 |
| Max. P | 0.999879 |

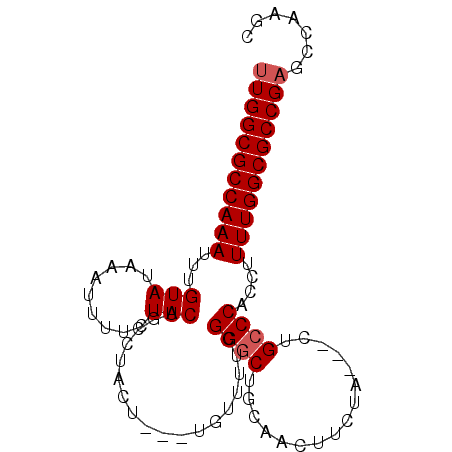
| Location | 4,353,754 – 4,353,844 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -21.47 |
| Energy contribution | -21.87 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
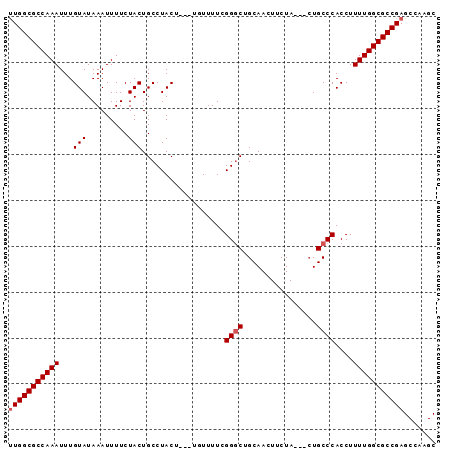
>3L_DroMel_CAF1 4353754 90 + 23771897 UUGGCGCCAAAUUUGUAUAAAUUUUCUACUGCCUACU---UUUAUUUGGGCUGCAACUUCUACUACUGCCCACCUUUUGGCGCCGAGCCAAGC (((((((((((...((..............(((((..---......))))).(((...........)))..))..)))))))))))....... ( -24.30) >DroSec_CAF1 4425 87 + 1 UUGGCGCCAAAUUUGUAUAAAUUUUCUACUGCCUACU---UGUUUUCGGGCUGCAACUUCUA---CUGCCCACCUUUUGGCGCCGAGCCAAGC (((((((((((...(((.........)))........---.......((((...........---..))))....)))))))))))....... ( -24.22) >DroSim_CAF1 4422 87 + 1 UUGGCGCCAAAUUUGUAUAAAUUUUCUACUGCCUACU---UGUUUUCGGGCUGCAACUUCUA---CUGCCCACCUUUUGGCGCCGAGCCAAGC (((((((((((...(((.........)))........---.......((((...........---..))))....)))))))))))....... ( -24.22) >DroEre_CAF1 4474 85 + 1 CUGGCGCCAAAUUUGUAUAAAUUUUCUACUGCCUACU-----UUUUCGGGCUGCAACUUCUA---CUGCCCACCUUUUGGCGCCGAGCCAAGC .((((((((((...((..............((((...-----.....)))).(((.......---.)))..))..))))))))))........ ( -23.20) >DroYak_CAF1 4475 90 + 1 UUGGCGCCAAAUUUGUAUAAAUUUUCUACUGCCUACUAUUUUUUUUUGGCCCGCAACUUCUA---GUGCCCACCUUUUGGCGCCGAGCCAAGC (((((((((((...((..............(((..............)))..(((.......---.)))..))..)))))))))))....... ( -22.44) >consensus UUGGCGCCAAAUUUGUAUAAAUUUUCUACUGCCUACU___UGUUUUCGGGCUGCAACUUCUA___CUGCCCACCUUUUGGCGCCGAGCCAAGC (((((((((((...(((.........)))..................((((................))))....)))))))))))....... (-21.47 = -21.87 + 0.40)

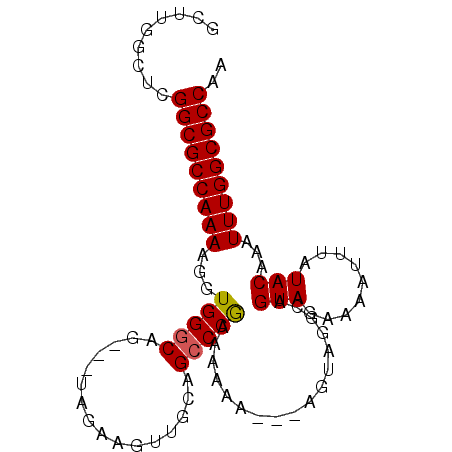
| Location | 4,353,754 – 4,353,844 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.11 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
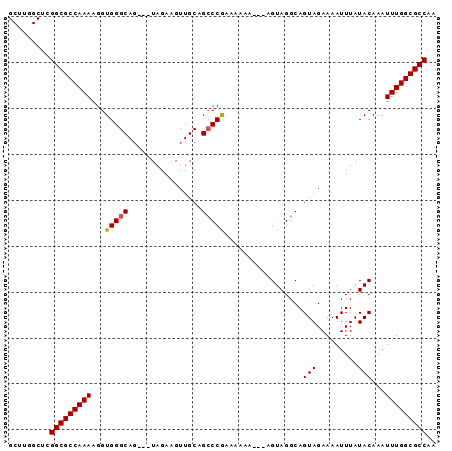
>3L_DroMel_CAF1 4353754 90 - 23771897 GCUUGGCUCGGCGCCAAAAGGUGGGCAGUAGUAGAAGUUGCAGCCCAAAUAAA---AGUAGGCAGUAGAAAAUUUAUACAAAUUUGGCGCCAA .........(((((((((...(((((.((((......)))).)))))......---........(((.........)))...))))))))).. ( -28.40) >DroSec_CAF1 4425 87 - 1 GCUUGGCUCGGCGCCAAAAGGUGGGCAG---UAGAAGUUGCAGCCCGAAAACA---AGUAGGCAGUAGAAAAUUUAUACAAAUUUGGCGCCAA .........(((((((((...(((((.(---(((...)))).)))))......---........(((.........)))...))))))))).. ( -26.50) >DroSim_CAF1 4422 87 - 1 GCUUGGCUCGGCGCCAAAAGGUGGGCAG---UAGAAGUUGCAGCCCGAAAACA---AGUAGGCAGUAGAAAAUUUAUACAAAUUUGGCGCCAA .........(((((((((...(((((.(---(((...)))).)))))......---........(((.........)))...))))))))).. ( -26.50) >DroEre_CAF1 4474 85 - 1 GCUUGGCUCGGCGCCAAAAGGUGGGCAG---UAGAAGUUGCAGCCCGAAAA-----AGUAGGCAGUAGAAAAUUUAUACAAAUUUGGCGCCAG .........(((((((((...(((((.(---(((...)))).)))))....-----........(((.........)))...))))))))).. ( -26.50) >DroYak_CAF1 4475 90 - 1 GCUUGGCUCGGCGCCAAAAGGUGGGCAC---UAGAAGUUGCGGGCCAAAAAAAAAUAGUAGGCAGUAGAAAAUUUAUACAAAUUUGGCGCCAA ..(((((((..((((....)))).(((.---.......))))))))))............(((.((...((((((....)))))).))))).. ( -24.70) >consensus GCUUGGCUCGGCGCCAAAAGGUGGGCAG___UAGAAGUUGCAGCCCGAAAAAA___AGUAGGCAGUAGAAAAUUUAUACAAAUUUGGCGCCAA .........(((((((((...(((((................))))).................(((.........)))...))))))))).. (-23.15 = -23.11 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:35 2006