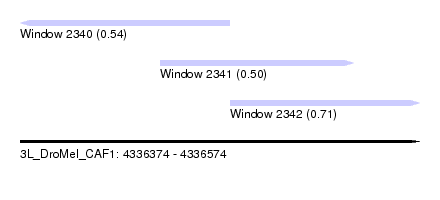
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,336,374 – 4,336,574 |
| Length | 200 |
| Max. P | 0.707322 |

| Location | 4,336,374 – 4,336,479 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.90 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4336374 105 - 23771897 CCUCCACAU-----GGAGGAGCUAUGGCGGAAAUCGUUAAAUA-----CUCGG-----GCGAUAAGACUGAAAGUGUUGCCGCAACAUGUUGCAGGGGCGAAGUCAUUGAACCGCAGGAA (((((....-----))))).......((((..((((((.....-----....)-----)))))..((((.....((((.(.((((....)))).).)))).))))......))))..... ( -32.50) >DroSec_CAF1 22716 104 - 1 CCUCCACAU-----GGAGGAGCUAUGGCGGAAAUCGUUAAAUA-----CUUUG-----GCGAUAAGACUGAAAGUGUUGCCGCAACAUGUUGCUG-AGCGAAGUCAUUGAACCGCAGGAA (((((....-----))))).......((((..((((((((...-----..)))-----)))))..((((.....((((.(.((((....)))).)-)))).))))......))))..... ( -32.60) >DroSim_CAF1 22731 104 - 1 CCUCCACAU-----GGAGGAGCUAUGGCGGAAAUCGUUAAAUA-----CUCUG-----GCGAUAAGACUGAAAGUGUUGCCGCAACAUGUUGCUG-GGCGAAGUCAUUGAACCGCAGGAA (((((....-----))))).......((((..(((((((....-----...))-----)))))..((((.......(((((((((....))))..-)))))))))......))))..... ( -33.11) >DroYak_CAF1 22164 104 - 1 CCUCCACAU-----GGAGGAGCUAUGGCGGAAAUCGUUAAAUA-----CUCUG-----GCGAUAAGACUGGAAAUGUUGCCGCAACAUGUUGCUG-GGCGAAGUCAUUGAACCGCAGGAA (((((....-----))))).......((((..(((((((....-----...))-----)))))..((((.......(((((((((....))))..-)))))))))......))))..... ( -33.11) >DroAna_CAF1 25345 113 - 1 CCUCCACAUGAAGUGGAGGAACUAUGGCGGAAAUCAUUAAAAAAAGCAAGCUGGAAAUGCGAUAAGAGUCAAGAUGUUGCUGCAACAUGUUGCAG-------GUCAUUGAACCGGCACUG (((((((.....))))))).......((.................))..(((((......(((....)))..((((...((((((....))))))-------..))))...))))).... ( -32.33) >consensus CCUCCACAU_____GGAGGAGCUAUGGCGGAAAUCGUUAAAUA_____CUCUG_____GCGAUAAGACUGAAAGUGUUGCCGCAACAUGUUGCUG_GGCGAAGUCAUUGAACCGCAGGAA (((((.........))))).......((((..(((((.....................)))))..((((.....((((.(.((((....)))).).)))).))))......))))..... (-21.42 = -22.90 + 1.48)

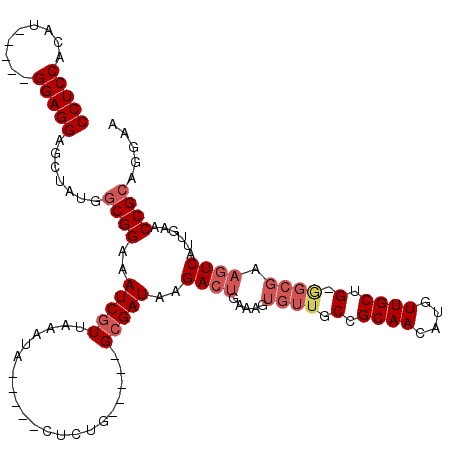
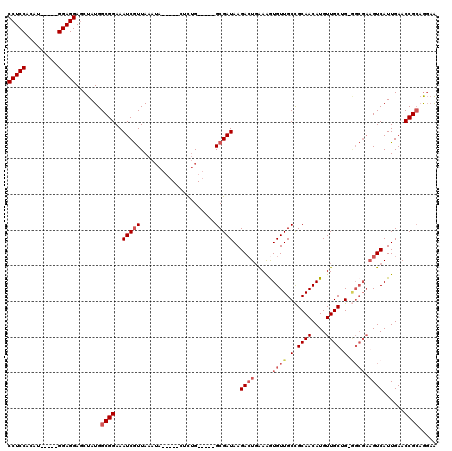
| Location | 4,336,444 – 4,336,541 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -12.71 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4336444 97 + 23771897 UUAACGAUUUCCGCCAUAGCUCCUCCAUGUGGAGGAG-UUUUUAACUUGCCCACACA-------AUG---------CAAAAAAGUAAAAAAAAAAAAUACAAUAAAGUUUGUUC ..(((((..........(((((((((....)))))))-))......((((.......-------..)---------))).............................))))). ( -17.20) >DroSec_CAF1 22785 103 + 1 UUAACGAUUUCCGCCAUAGCUCCUCCAUGUGGAGGAG-UU-CCAACUUGCCCACACA-------AAAAAAAAAAUCUGAAAAAGUUAA--GAAAAAAGGCAACAAAGUUUGUUC ............(((..(((((((((....)))))))-))-..(((((.........-------.................)))))..--.......))).............. ( -21.17) >DroSim_CAF1 22800 110 + 1 UUAACGAUUUCCGCCAUAGCUCCUCCAUGUGGAGGAG-UU-CCAACUUGCCCACACACCAAAAAAAAAAAAAAAUCCAAAAAAGUAGA--GAAAAAAUACAACAAAGUUUGUUC ..(((((..........(((((((((....)))))))-))-..........................................(((..--.......)))........))))). ( -16.10) >DroYak_CAF1 22233 103 + 1 UUAACGAUUUCCGCCAUAGCUCCUCCAUGUGGAGGAGGUU-CCAGCUUGCCCACACA-------CAAAUAAAAGUAUGAAAAAA-AAA--CAACAAAGACAACAUAGUUUGUUU ............((...(.(((((((....))))))).).-...)).......((.(-------(........)).))......-...--......((((((......)))))) ( -16.80) >consensus UUAACGAUUUCCGCCAUAGCUCCUCCAUGUGGAGGAG_UU_CCAACUUGCCCACACA_______AAAAAAAAAAUCCAAAAAAGUAAA__GAAAAAAGACAACAAAGUUUGUUC ...................(((((((....))))))).............................................................((((......)))).. (-12.71 = -12.52 + -0.19)


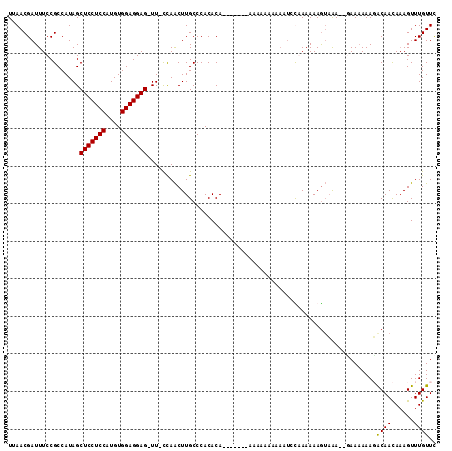
| Location | 4,336,479 – 4,336,574 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -14.95 |
| Consensus MFE | -8.06 |
| Energy contribution | -8.75 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4336479 95 + 23771897 AG-UUUUUAACUUGCCCACACA-------AUG---------CAAAAAAGUAAAAAAAAAAAAUACAAUAAAGUUUGUUCGAAUU-UUUGCAACAAACAAGAUGGCCAAGCAAA ..-.......((((.((((...-------.((---------(......)))....................((((((((((...-.))).)))))))..).))).)))).... ( -14.00) >DroSec_CAF1 22820 101 + 1 AG-UU-CCAACUUGCCCACACA-------AAAAAAAAAAUCUGAAAAAGUUAA--GAAAAAAGGCAACAAAGUUUGUUCGAAUU-UUUGCAACAAACAGCAUGGCCAAGCAAG ..-..-....(((((.......-------..........(((..........)--)).....(((......((((((((((...-.))).)))))))......)))..))))) ( -17.60) >DroSim_CAF1 22835 108 + 1 AG-UU-CCAACUUGCCCACACACCAAAAAAAAAAAAAAAUCCAAAAAAGUAGA--GAAAAAAUACAACAAAGUUUGUUCGAAUU-UUUGCAACAAACAGCAUGGCCAAGCAAG ..-..-....((((.(((..(...........................(((..--.......)))......((((((((((...-.))).))))))).)..))).)))).... ( -13.90) >DroYak_CAF1 22268 97 + 1 AGGUU-CCAGCUUGCCCACACA-------CAAAUAAAAGUAUGAAAAAA-AAA--CAACAAAGACAACAUAGUUUGUUUGAAUUUUUUGCAACAAACAAGAUGG-----CAAG .....-....((((((......-------((((((((..((((......-...--............)))).))))))))....(((((.......))))).))-----)))) ( -14.30) >consensus AG_UU_CCAACUUGCCCACACA_______AAAAAAAAAAUCCAAAAAAGUAAA__GAAAAAAGACAACAAAGUUUGUUCGAAUU_UUUGCAACAAACAACAUGGCCAAGCAAG ..........((((((((..............................(((...........)))......((((((((((.....))).)))))))....)))....))))) ( -8.06 = -8.75 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:22 2006