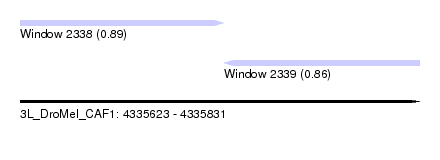
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,335,623 – 4,335,831 |
| Length | 208 |
| Max. P | 0.887544 |

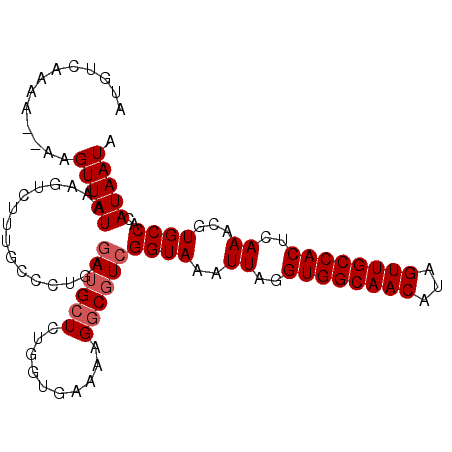
| Location | 4,335,623 – 4,335,729 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -23.11 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4335623 106 + 23771897 CCACUCGAAUCCACUGUA----------CUCCAUCUCCAUUACUCAUGGAGAGCCCGGGCCCAGCCCAUUCCCCGUUUAGCAACGUUUUAUGGCGGCGAACUGCAGUUUUUAAUAA ............((((((----------.....(((((((.....)))))))(((.((((...)))).....((((..(((...)))..)))).)))....))))))......... ( -28.30) >DroSec_CAF1 22037 105 + 1 CUACUCGAAUCCACUGUA----------CUCCGUCUCCAUUACUCAUGGAGAGCCCG-GCCCAGCCCAUUCUCCGUUUAGCAACGUUUUAUGGCGGCGAACUGCAGUUUUUAAUAA ............((((((----------...((((.((((.((....((((((...(-((...)))..))))))(((....)))))...)))).))))...))))))......... ( -26.50) >DroSim_CAF1 22008 106 + 1 CCACUCGAAUCCACUGUA----------CUCCGUCUCCAUUACUCAUGGAGAGCCAGGGCCCAGCCCAUUCCCCGUUUAGCAACGUUUUAUGGCGGCGAACUGCAGUUUUUAAUAA ............((((((----------(.((.(((((((.....)))))))((((((((...))))......((((....)))).....)))))).)...))))))......... ( -29.00) >DroYak_CAF1 21359 116 + 1 CCACUCCAAUCCACCGUACUACUCCAUACUCCAUCUCCAUUACUCAUGGAGAGGCCAGGCCCAACCCAUUCCCCGUUUAGCAACGUUUUAUGGCGGUGAACUGCGGUUUUUAAUAA ............((((((...(.((........(((((((.....))))))).((((((......))......((((....)))).....)))))).)...))))))......... ( -26.00) >consensus CCACUCGAAUCCACUGUA__________CUCCAUCUCCAUUACUCAUGGAGAGCCCGGGCCCAGCCCAUUCCCCGUUUAGCAACGUUUUAUGGCGGCGAACUGCAGUUUUUAAUAA ............((((((...............(((((((.....)))))))......(((....((((....((((....))))....)))).)))....))))))......... (-23.11 = -22.55 + -0.56)

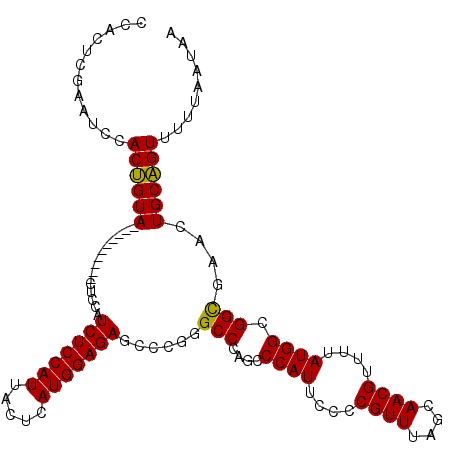
| Location | 4,335,729 – 4,335,831 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.45 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4335729 102 - 23771897 ACGUCAAAA--AAGUUAUAAGUUCUUUGCCCUGAGAUGCUCUUAUGAAAAGGCGUCGGUAAAUUAGGUGGCAACAUAGUUGCCACUCAAACGUGCCACAUAAUA (((((....--...(((((((.((((......))))....)))))))...))))).((((..((..((((((((...))))))))..))...))))........ ( -28.70) >DroSec_CAF1 22142 99 - 1 AUGUCAAAA--AAGUUAUAAAGU---UGCCCUGCUAUGCUCUGGUGAAAAGGCGUCGGUAAAUUAGGUGGCAACAUAGUUGCCACUCAAACGUGCCACAUAAUA .........--..(((((....(---..((..((...))...))..)...((((.((.....((..((((((((...))))))))..)).))))))..))))). ( -24.50) >DroSim_CAF1 22114 102 - 1 AUGUCAAAA--GAGUUAUAAAGUCUUUGCCCUGCGAUGCUCUGGUGAAAAGGCGUCGGUAAAUUAGGUGGCAACAUAGUUGCCACUCAAACGUGCCACAUAAUA ...(((..(--((((.....((........)).....)))))..)))...((((.((.....((..((((((((...))))))))..)).))))))........ ( -28.60) >DroYak_CAF1 21475 103 - 1 AUGCCAAAAUAUAGUUAUAAAGUCUUUGCCCCAAGAUGCUCGUCUGAAAAG-CCUCGGUAAAUUAGGUGGCAACAUAGUUGCCACUCAAACGUGCCACAUAAUA .............(((((..((.((((......((((....))))..))))-.)).((((..((..((((((((...))))))))..))...))))..))))). ( -22.90) >consensus AUGUCAAAA__AAGUUAUAAAGUCUUUGCCCUGAGAUGCUCUGGUGAAAAGGCGUCGGUAAAUUAGGUGGCAACAUAGUUGCCACUCAAACGUGCCACAUAAUA .............(((((................((((((..........))))))((((..((..((((((((...))))))))..))...))))..))))). (-21.70 = -22.45 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:19 2006