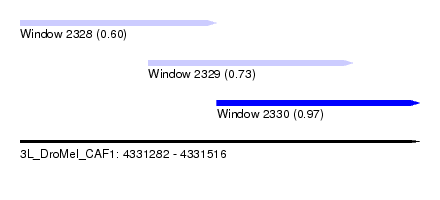
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,331,282 – 4,331,516 |
| Length | 234 |
| Max. P | 0.974906 |

| Location | 4,331,282 – 4,331,397 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -15.15 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4331282 115 + 23771897 --U-UAAAUGCCAGCAAAAGAGGAAAUGUUUACAGCACAGCCAA--CUGCAAUCCAUUAGCUAUUUUUAUAGCAGCACAAAAGCACAAAAAUAAUAAACAAAAUCUACACAUAUUUCUUU --.-...............((((((((((.....(((.......--.))).....(((((((((....))))).((......)).......))))...............)))))))))) ( -15.00) >DroSec_CAF1 17790 114 + 1 --U-UAAAUGCCAGCAA-AGAGGAAAUGUUUACAGCACAGCCAA--CUGCAAUCCAUUAGCUAUUUUUAUAGCAGCACAAAAGCACAAAAAUAACAAACAAAAUCUACACAUAUUUCUUA --.-.............-..(((((((((.....(((.......--.))).........(((((....))))).((......))..........................))))))))). ( -14.30) >DroSim_CAF1 16384 115 + 1 --UUUAUAUGCCAGCAA-AGAGGAAAUGUUUACAGCACAGCCAA--CUGCAAUCCAUUAGCUAUUUUUAUAGCAGCACAAAAGCACAAAAAUAACAAACAAAAUCUACACAUAUUUCUUA --...............-..(((((((((.....(((.......--.))).........(((((....))))).((......))..........................))))))))). ( -14.30) >DroEre_CAF1 17778 108 + 1 UUU-UAAAUGCCAGCAA-AGAGGAAAUGUUUACAGCACUGCCAA--CUGCAAUCCAUUAGCUAUUUUUAUAGCAGCAC--------AGAAAUAACAAACAAAAUCUACACAUAUUUCUUU ...-....(((..(((.-...((...(((.....)))...))..--.))).........(((((....))))).))).--------(((((((..................))))))).. ( -14.27) >DroYak_CAF1 16963 108 + 1 --U-GAAAUGCCAGCAA-AGAGGAAAUGUUUACAGCACUGCCAACACUGCAAUGCAUUAGCUAUUUUUAUAGCAGCAC--------AAAAAUAACAAACAAAAUCUACACAUAUUUCUUU --.-.............-.((((((((((.....(((.(((.......))).)))....(((((....))))).....--------........................)))))))))) ( -17.90) >consensus __U_UAAAUGCCAGCAA_AGAGGAAAUGUUUACAGCACAGCCAA__CUGCAAUCCAUUAGCUAUUUUUAUAGCAGCACAAAAGCACAAAAAUAACAAACAAAAUCUACACAUAUUUCUUU ........(((..(((.....((...(((.....)))...)).....))).........(((((....))))).)))........................................... (-12.70 = -12.70 + 0.00)

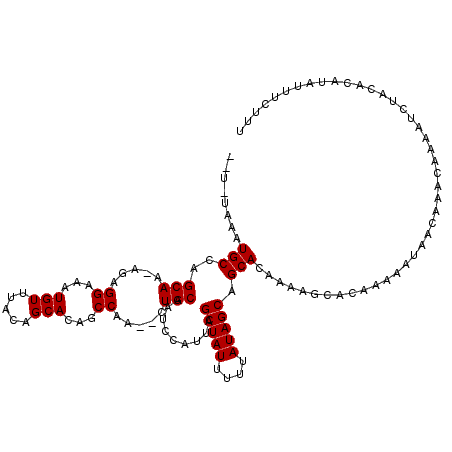
| Location | 4,331,357 – 4,331,477 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -23.39 |
| Consensus MFE | -20.56 |
| Energy contribution | -19.92 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4331357 120 + 23771897 AAGCACAAAAAUAAUAAACAAAAUCUACACAUAUUUCUUUGAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAGAGGGGAAAUGGUGAGUAUAAAACGUAGACAC .......................(((((...(((((((......)))))))(..(((((((((((....)))..((.((....)))).....))))))))..).........)))))... ( -25.20) >DroSec_CAF1 17864 120 + 1 AAGCACAAAAAUAACAAACAAAAUCUACACAUAUUUCUUAGAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGGAAAUGCAGGGGGGAAAUGGCGGGUAUAAAACGCAGACAC ..((...........................(((((((......)))))))((((((((((((((....)))..((.((....)))).....))))))))))).........))...... ( -23.40) >DroSim_CAF1 16459 120 + 1 AAGCACAAAAAUAACAAACAAAAUCUACACAUAUUUCUUAGAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGGAAAUGCAGGGGGGAAAUGGCGGGUAUAAAACGCAGACAC ..((...........................(((((((......)))))))((((((((((((((....)))..((.((....)))).....))))))))))).........))...... ( -23.40) >DroEre_CAF1 17852 113 + 1 ------AGAAAUAACAAACAAAAUCUACACAUAUUUCUUUGAGCGGGCAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAA-GGGGAAAUGGCGAGUAUAAAACGAAGACAC ------.............................((((((.(....)...(((((((((((.(((.((((((...)))))).)))...-..)))))))))))........))))))... ( -22.50) >DroYak_CAF1 17037 113 + 1 ------AAAAAUAACAAACAAAAUCUACACAUAUUUCUUUGAGCGCGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAA-GGGGAAGUGGCGAGUAUAAAACGAAGACAC ------.............................((((((..........(((((((((((.(((.((((((...)))))).)))...-..)))))))))))........))))))... ( -22.47) >consensus AAGCACAAAAAUAACAAACAAAAUCUACACAUAUUUCUUUGAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAG_GGGGAAAUGGCGAGUAUAAAACGAAGACAC .......................(((.(...(((((((......)))))))(((((((((((.(((.((((((...)))))).)))......))))))))))).........).)))... (-20.56 = -19.92 + -0.64)

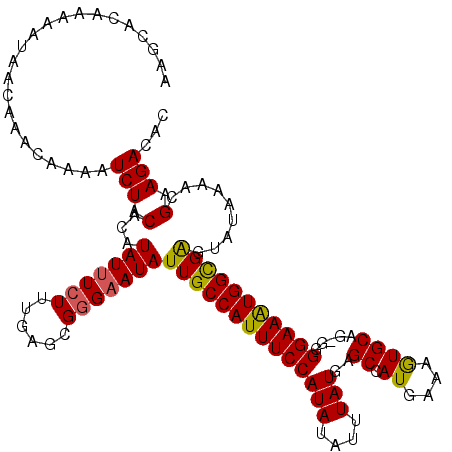
| Location | 4,331,397 – 4,331,516 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -32.44 |
| Energy contribution | -31.56 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
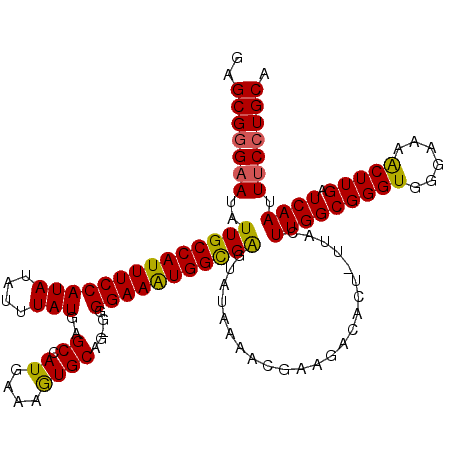
>3L_DroMel_CAF1 4331397 119 + 23771897 GAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAGAGGGGAAAUGGUGAGUAUAAAACGUAGACACU-UUACUUGGCGGGUGGGAAAACUUGAUCAAUUUCCUGCA ..(((((((..(..(((((((((((....)))..((.((....)))).....))))))))..).........(((((...)-))))(((((((((......))))).)))).))))))). ( -33.20) >DroSec_CAF1 17904 120 + 1 GAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGGAAAUGCAGGGGGGAAAUGGCGGGUAUAAAACGCAGACACUUUUACUUGGCGGGUGGGAAAACUUGAUCAAUUUCCUGCA ..(((((((..((((((((((((((....)))..((.((....)))).....))))))))))).......................(((((((((......))))).)))).))))))). ( -34.10) >DroSim_CAF1 16499 120 + 1 GAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGGAAAUGCAGGGGGGAAAUGGCGGGUAUAAAACGCAGACACUUUUACUUGGCGGGUGGGAAAACUUGAUCAAUUUCCUGCA ..(((((((..((((((((((((((....)))..((.((....)))).....))))))))))).......................(((((((((......))))).)))).))))))). ( -34.10) >DroEre_CAF1 17886 118 + 1 GAGCGGGCAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAA-GGGGAAAUGGCGAGUAUAAAACGAAGACACU-UUAGUUGGCGGGUGGGAAAGCUUGAUCAAUUUCCUGCA ..(((((....(((((((((((.(((.((((((...)))))).)))...-..)))))))))))..................-..(((((((((((......))))).)))))).))))). ( -33.40) >DroYak_CAF1 17071 118 + 1 GAGCGCGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAA-GGGGAAGUGGCGAGUAUAAAACGAAGACACU-UUAGUUGGCGGGUGGGAAAGCUUGAUCAAUUUCCUGCA ...(((.(((.(((((((((((.(((.((((((...)))))).)))...-..))))))))))).........((((...))-)).))).))).(..(((((..........)))))..). ( -32.30) >consensus GAGCGGGAAUAUUGCCAUUUCCAUAUAUUUAUGAGCCAUGAAAGUGCAG_GGGGAAAUGGCGAGUAUAAAACGAAGACACU_UUACUUGGCGGGUGGGAAAACUUGAUCAAUUUCCUGCA ..(((((((..(((((((((((.(((.((((((...)))))).)))......))))))))))).......................(((((((((......))))).)))).))))))). (-32.44 = -31.56 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:10 2006