
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,330,676 – 4,330,897 |
| Length | 221 |
| Max. P | 0.996344 |

| Location | 4,330,676 – 4,330,788 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -30.20 |
| Energy contribution | -29.88 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4330676 112 + 23771897 CCCAGAAC--CAGCGAUUGGAUAAAGCUGCCCCGCUCUAUAAAUAGAGCCCUGGUCGAGCCUUAUCAGCCUCGAUCCGCGUGGCCAGAGUUUCGCUUUUCAAGUCUCU-GGCUUU .......(--(((.((((.((.(((((......((((((....)))))).(((((((.((...(((......)))..)).)))))))......))))))).)))).))-)).... ( -35.70) >DroSec_CAF1 17179 112 + 1 CCCAGAAC--CAGCGAUUGGAUAAAGCUGCUCCGCUCUAUAAAUAGAGCCCUGGUCGCGCCUUAUCAGCCUCGAUCCGCGUGGCCAGAGUUUCGCUUUUCAAGUCUCU-GGCUUU .......(--(((.((((.((.(((((.((((.((((((....))))))..(((((((((...(((......)))..)))))))))))))...))))))).)))).))-)).... ( -42.40) >DroSim_CAF1 15771 112 + 1 CCCAGAAC--CAGCGAUUGGAUAAAGCUGCCCCGCUCUAUAAAUAGAGCCCUGGCCGAGCCUUAUCAGCCUCGAUCCGCGUGGCCAGAGUUUCGCUUUUCAAGUCUCU-GGCUUU .......(--(((.((((.((.(((((......((((((....)))))).(((((((.((...(((......)))..)).)))))))......))))))).)))).))-)).... ( -38.40) >DroEre_CAF1 17187 112 + 1 CCCAGAAC--CACCGAUUGGAUGAAGCUGCC-CGCUCUAUAAAUAGAGCCCUGGUCGGGCCUUAUCAGCCUCGAUCCGCGUGGCCAGAGUUUCGCUUUUCAAGUCGCUUUGUUUU ...(((((--...(((((((((((((((...-.((((((....)))))).(((((((.((...(((......)))..)).))))))))))))))...))).)))))....))))) ( -34.10) >DroYak_CAF1 16370 114 + 1 CCCAGAACAGCAGCGAUUGGAUAAAGCUGCCCCGCUCUAUAAAUAGAGCCCUGGUCGCGCCUUAUCAGCCUCGAUCCGCGUGGCCAGAGUUUCGCUUUUCAAGUCUCU-GGCUUU .(((((((.(((((...........)))))...((((((....)))))).((((((((((...(((......)))..))))))))))...............)).)))-)).... ( -40.70) >consensus CCCAGAAC__CAGCGAUUGGAUAAAGCUGCCCCGCUCUAUAAAUAGAGCCCUGGUCGAGCCUUAUCAGCCUCGAUCCGCGUGGCCAGAGUUUCGCUUUUCAAGUCUCU_GGCUUU ..............((((.((.(((((......((((((....)))))).(((((((.((...(((......)))..)).)))))))......))))))).)))).......... (-30.20 = -29.88 + -0.32)

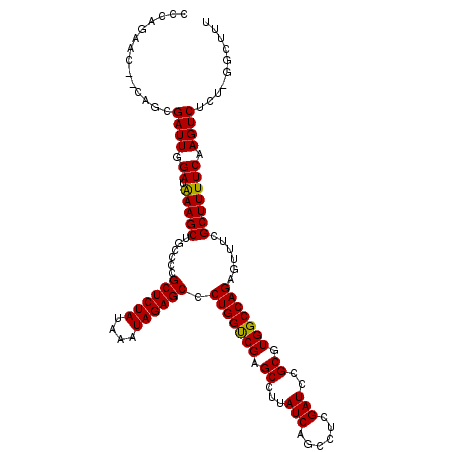

| Location | 4,330,676 – 4,330,788 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -31.56 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4330676 112 - 23771897 AAAGCC-AGAGACUUGAAAAGCGAAACUCUGGCCACGCGGAUCGAGGCUGAUAAGGCUCGACCAGGGCUCUAUUUAUAGAGCGGGGCAGCUUUAUCCAAUCGCUG--GUUCUGGG ....((-((((.((.....(((((......((((.((.....)).))))(((((((((.(.((...((((((....))))))..))))))))))))...))))))--))))))). ( -40.90) >DroSec_CAF1 17179 112 - 1 AAAGCC-AGAGACUUGAAAAGCGAAACUCUGGCCACGCGGAUCGAGGCUGAUAAGGCGCGACCAGGGCUCUAUUUAUAGAGCGGAGCAGCUUUAUCCAAUCGCUG--GUUCUGGG ....((-((((.((.....(((((......((((.((.....)).))))((((((((((..((...((((((....)))))))).)).))))))))...))))))--))))))). ( -39.70) >DroSim_CAF1 15771 112 - 1 AAAGCC-AGAGACUUGAAAAGCGAAACUCUGGCCACGCGGAUCGAGGCUGAUAAGGCUCGGCCAGGGCUCUAUUUAUAGAGCGGGGCAGCUUUAUCCAAUCGCUG--GUUCUGGG ....((-((((.((.....(((((......((((.((.....)).))))(((((((((..(((...((((((....))))))..))))))))))))...))))))--))))))). ( -43.10) >DroEre_CAF1 17187 112 - 1 AAAACAAAGCGACUUGAAAAGCGAAACUCUGGCCACGCGGAUCGAGGCUGAUAAGGCCCGACCAGGGCUCUAUUUAUAGAGCG-GGCAGCUUCAUCCAAUCGGUG--GUUCUGGG ....((.(((.(((.((...(((............)))((((.(((((((.......((.....))((((((....)))))).-..)))))))))))..))))).--))).)).. ( -34.40) >DroYak_CAF1 16370 114 - 1 AAAGCC-AGAGACUUGAAAAGCGAAACUCUGGCCACGCGGAUCGAGGCUGAUAAGGCGCGACCAGGGCUCUAUUUAUAGAGCGGGGCAGCUUUAUCCAAUCGCUGCUGUUCUGGG ...(((-((((..(((.....)))..)))))))..((((.((((....))))....)))).(((((((((((....))))))..((((((...........))))))..))))). ( -42.40) >consensus AAAGCC_AGAGACUUGAAAAGCGAAACUCUGGCCACGCGGAUCGAGGCUGAUAAGGCGCGACCAGGGCUCUAUUUAUAGAGCGGGGCAGCUUUAUCCAAUCGCUG__GUUCUGGG ............((.(((.(((((.......((...))((((.(((((((....((.....))...((((((....))))))....)))))))))))..)))))....))).)). (-31.56 = -31.60 + 0.04)


| Location | 4,330,788 – 4,330,897 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -21.66 |
| Consensus MFE | -12.73 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4330788 109 + 23771897 UUUCCUUUUCUUUUCUUUUUGU-------UUUUCACUCGCUCUCGAGUGGCCAUAAAAAUUUGUUGCAUUUCCGUUGGCUUGUACAUGAAAAUAUGCAGCAACUCUCCU----CUUGCAA ...............(((((((-------...(((((((....)))))))..))))))).(((((((((......((.......)).......))))))))).......----....... ( -20.42) >DroEre_CAF1 17299 104 + 1 UAUCCAUUUCUUUUCC-----------UUUUCUCGCUCGCUCUCGAGUGGCCAUAAAAAUUUGUUGCAUUUCCGUU-GCUUGUACAAGAAAAUAUGCAGCAACUCUCUUGCAACUU---- ................-----------.....(((((((....)))))))............((((((.....(((-(((.(((..........))))))))).....))))))..---- ( -22.80) >DroYak_CAF1 16484 111 + 1 UUUCCUUUUCUUUUCUUUUUU----CAUUUUUUCACUCGCUCUCGAGUGGCCAUAAAAAUUUGUUGCAUUUCCGUU-GCUUGUACAAGAAAAUAUGCAGCAACUCUCUUGCAACUU---- .....................----.......(((((((....)))))))............((((((.....(((-(((.(((..........))))))))).....))))))..---- ( -23.30) >DroAna_CAF1 16869 96 + 1 UUUCGCUUCAGCCUCCC-AGGCUAG-UCUUUUUCUCUCUCUGCGGAGUGGCCAUAAAAAUUUGUUGCAUUUCCGUU-GCUUGUACAACAAACUAUGCAG--------------------- ....(((..((((....-.))))))-)............((((((((..((.((((....)))).))..))))(((-(......)))).......))))--------------------- ( -20.10) >consensus UUUCCUUUUCUUUUCCU_UUG______UUUUUUCACUCGCUCUCGAGUGGCCAUAAAAAUUUGUUGCAUUUCCGUU_GCUUGUACAAGAAAAUAUGCAGCAACUCUCUU____CUU____ ................................(((((((....)))))))..........(((((((((........................))))))))).................. (-12.73 = -13.23 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:06 2006