| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,329,297 – 4,329,411 |
| Length | 114 |
| Max. P | 0.505182 |

| Location | 4,329,297 – 4,329,411 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
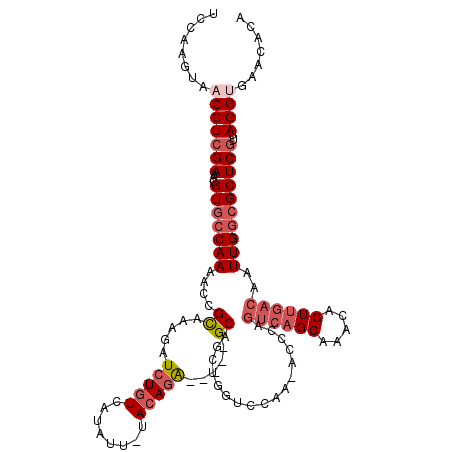
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -20.51 |
| Energy contribution | -22.07 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
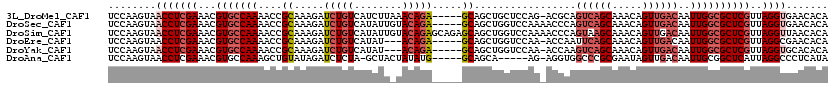
>3L_DroMel_CAF1 4329297 114 + 23771897 UCCAAGUAACCUCGAAACGUGCCAAAACCGCAAAGAUCUGUCAUCUUAAACAGA-----GCAGCUGCUCCAG-ACGCAGUCAGCAAACAGUUGACAAUUGGCGCUCGUUAGGUGAACACA .....((.(((((((...(((((((....((.(((((.....))))).....((-----((....))))...-..)).((((((.....))))))..))))))))))..))))..))... ( -33.60) >DroSec_CAF1 15871 115 + 1 UCCAAGUAACCUCGAAACGUGCCAAAACCGCAAAGAUCUGUCAUAUUGUACAGA-----GCAGCUGGUCCAAAACCCAGUCAGCAAACAGUUGACAAUUGGCGCUCGUUAGGUGAACACA .....((.(((((((...(((((((.(((((.....((((((.....).)))))-----...)).)))..........((((((.....))))))..))))))))))..))))..))... ( -32.30) >DroSim_CAF1 14436 120 + 1 UCCAAGUAACCUCGAAACGUGCCAAAACCGCAAAGAUCUGUCAUAUUGUACAGAGCAGAGCAGCUGGUCCAAAACCCAGUAAGCAAACAGUUGACAAUUGGCGCUCGUUAGGUUAACACA ......(((((((((...(((((((....((.....((((((.....).))))).....)).(((((........))))).................))))))))))..))))))..... ( -30.50) >DroEre_CAF1 15826 111 + 1 UCCAAGUAACCUCGAAACGUGCCAAAACCGCAAAGAUCUGUCAUAU---ACAGA-----GCAGCUGGUCCAA-ACCAAUUCAGCAAACAGUUGACAAUUGGCGCUCGUUAGGCGAACACA .....((..((((((...(((((((.(((((.....(((((.....---)))))-----...)).)))....-......(((((.....)))))...))))))))))..)))...))... ( -25.30) >DroYak_CAF1 14985 111 + 1 UCCAAGUAACCUCGAAACGUGCCAAAACCGCAAAGAUCUGUCAUAU---ACAGA-----GCAGCUGGUCCAA-ACCAAGUCAGCAAACAGUUGACAAUUGGCGCUCGUUAGGUGCACACA .....((.(((((((...(((((((.(((((.....(((((.....---)))))-----...)).)))....-.....((((((.....))))))..))))))))))..))))..))... ( -32.30) >DroAna_CAF1 15786 108 + 1 UCCAAGUAACCUCGAAACGUGCCAAAGCUGUAUAGAUCUCUA-GCUACUAUAUG-----GCAGCA-----AG-AGGUGGCCCGCGAAUAGUUGACAAUUGCGGCUCAUUAGGCCCUCAUA ........(((((......(((....(((((((((.......-....)))))))-----)).)))-----.)-))))((((....((((((((.(....)))))).))).))))...... ( -26.10) >consensus UCCAAGUAACCUCGAAACGUGCCAAAACCGCAAAGAUCUGUCAUAUU_UACAGA_____GCAGCUGGUCCAA_ACCCAGUCAGCAAACAGUUGACAAUUGGCGCUCGUUAGGUGAACACA ........(((((((...(((((((....((.....(((((........))))).....)).................((((((.....))))))..))))))))))..))))....... (-20.51 = -22.07 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:02 2006