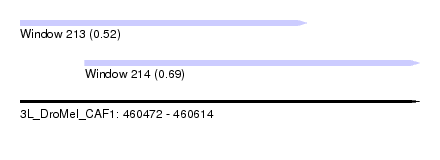
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 460,472 – 460,614 |
| Length | 142 |
| Max. P | 0.688762 |

| Location | 460,472 – 460,574 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.02 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 460472 102 + 23771897 AGGCCAAAUGUCAGUUUUCAUUGGCAAAGACCUGCAAUUUUUAGGGCUUCUCAUAGAG-GAACGCCUUCAAGAUAGCUUGAGCUGCCAGCCAUUUCUUUCACC .(((..((((........))))((((.......((.(((((.(((((((((......)-))).))))).))))).))......)))).)))............ ( -25.12) >DroSec_CAF1 41473 102 + 1 AGGCCAAAUGUCAGUUUUCAUUGGCAAAGACCUGCAAUUUUUAGGGCUUCUCAUAGAG-GAACGCCUUCAAGAUAGCUUGAGCUGCCAGCCAUUUCUUUCACC .(((..((((........))))((((.......((.(((((.(((((((((......)-))).))))).))))).))......)))).)))............ ( -25.12) >DroEre_CAF1 43405 102 + 1 AGGCCAAAUGUCAGUUUUCAUUGGCAGAGACCUGCAAUUUUUAGGGCUUCUCAUAGAA-GAACGCCUUCGAGAUAGUUUGGGCUGCCAGCAAUUUCUUUUGCC .(((.....))).........((((((...((.((.(((((.(((((((((......)-))).))))).))))).))..)).))))))((((......)))). ( -30.40) >DroYak_CAF1 42975 92 + 1 AGGCCAAAUGUCAGUUUUCAUUGGCAGAGACCUGCAAUUUCUAGGGCUUCUUAUAGAAGGAACGCCUUCGAGAUAGUU-GAGC----------UCUUUUAACC ..(((((.((........)))))))((((..(.((.(((((.((((((((((.....))))).))))).))))).)).-)..)----------)))....... ( -26.60) >consensus AGGCCAAAUGUCAGUUUUCAUUGGCAAAGACCUGCAAUUUUUAGGGCUUCUCAUAGAA_GAACGCCUUCAAGAUAGCUUGAGCUGCCAGCCAUUUCUUUCACC .((((((.(((((((....))))))).......((.(((((.((((((((.........))).))))).))))).))))).)))................... (-19.21 = -19.02 + -0.19)

| Location | 460,495 – 460,614 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -18.05 |
| Energy contribution | -20.68 |
| Covariance contribution | 2.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 460495 119 + 23771897 GCAAAGACCUGCAAUUUUUAGGGCUUCUCAUAGAG-GAACGCCUUCAAGAUAGCUUGAGCUGCCAGCCAUUUCUUUCACCAAAGUAGGCUGGUCACAACGGCUUGCAUUCAAAUCCCAUU ((((.(.((.((.(((((.(((((((((......)-))).))))).))))).))....(..(((((((....((((....))))..)))))))..)...))))))).............. ( -32.90) >DroSec_CAF1 41496 119 + 1 GCAAAGACCUGCAAUUUUUAGGGCUUCUCAUAGAG-GAACGCCUUCAAGAUAGCUUGAGCUGCCAGCCAUUUCUUUCACCAAAGUUGGCUGGUCGCAACGGCUUGCAUUCAAAUCCCAUU ((((.(.((.((.(((((.(((((((((......)-))).))))).))))).))....((.((((((((...((((....)))).)))))))).))...))))))).............. ( -39.20) >DroEre_CAF1 43428 118 + 1 GCAGAGACCUGCAAUUUUUAGGGCUUCUCAUAGAA-GAACGCCUUCGAGAUAGUUUGGGCUGCCAGCAAUUUCUUUUGCCAAAGUAGGCUGGUCGCAA-GCCUUCCACUCAAAUUCCAUU ((((....)))).(((((.(((((((((......)-))).))))).)))))(((..(((((((((((.....((((....))))...))))))....)-))))...)))........... ( -33.90) >DroYak_CAF1 42998 95 + 1 GCAGAGACCUGCAAUUUCUAGGGCUUCUUAUAGAAGGAACGCCUUCGAGAUAGUU-GAGC----------UCUUUUAACCAAAGAAGGCCGAUU--------------CCAAAUUCCAUU ((((....)))).(((((.((((((((((.....))))).))))).)))))((((-(.((----------.(((((......))))))))))))--------------............ ( -25.50) >consensus GCAAAGACCUGCAAUUUUUAGGGCUUCUCAUAGAA_GAACGCCUUCAAGAUAGCUUGAGCUGCCAGCCAUUUCUUUCACCAAAGUAGGCUGGUCGCAA_GGCUUGCAUUCAAAUCCCAUU ((((....)))).(((((.((((((((.........))).))))).))))).......((.(((((((....((((....))))..))))))).))........................ (-18.05 = -20.68 + 2.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:53 2006