
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,328,756 – 4,328,903 |
| Length | 147 |
| Max. P | 0.965674 |

| Location | 4,328,756 – 4,328,875 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.80 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -28.59 |
| Energy contribution | -28.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
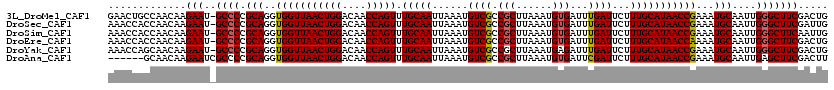
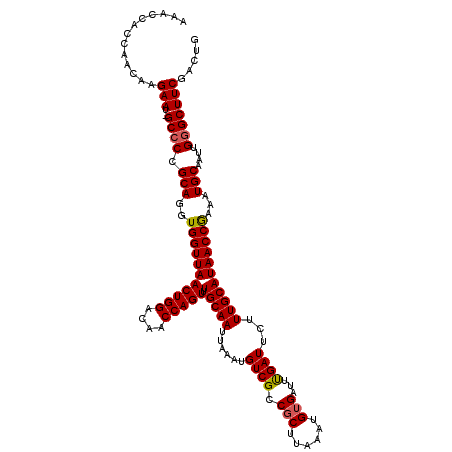
>3L_DroMel_CAF1 4328756 119 - 23771897 GAACUGCCAACAAGAAU-GCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUUUGAUUCUUUGCAUAACCGAAAUGCAAUUGGGCUUCGACUG .............(((.-((((.(((..(((((((((((....))))).(((((......((((.(((......)))...))))...)))))))))))...)))....)))))))..... ( -31.60) >DroSec_CAF1 15327 119 - 1 AAACCACCAACAAGAAU-GCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUUUGAUUCUUUGCAUAACCGAAAUGCAAUUGGGCUUCGAUUG .............(((.-((((.(((..(((((((((((....))))).(((((......((((.(((......)))...))))...)))))))))))...)))....)))))))..... ( -31.60) >DroSim_CAF1 13888 119 - 1 AAACCACCAACAAGAAU-GCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUUUGAUUCUUUGCAUAACCGAAAUGCAAUUGGGCUUCAAUUG .............(((.-((((.(((..(((((((((((....))))).(((((......((((.(((......)))...))))...)))))))))))...)))....)))))))..... ( -31.60) >DroEre_CAF1 15286 119 - 1 AAACCACCAACAAGAAU-GCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUUUGAUUCUUUGCAUAACCAAAAUGCAAUUGGGCUUCGACUG .............(((.-((((.(((..(((((((((((....))))).(((((......((((.(((......)))...))))...)))))))))))...)))....)))))))..... ( -32.30) >DroYak_CAF1 14431 119 - 1 AAACCAGCAACAAGAAU-GCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGAGAUUUGAUUCUUUGCAUAACCGAAAUGCAAUUGGGCUUCGACUG .............(((.-((((.(((..(((((((((((....))))).(((((((((((.(((.........))).)))))))....))))))))))...)))....)))))))..... ( -31.90) >DroAna_CAF1 15280 114 - 1 ------GCAACAAGAAUCGCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUUCGAUUCUUUGCAUAACCGAAAUGCAAUUGAGCUUCGACUU ------(((.....(((((((.....)))))))((((((....)))))))))........((((..((((((.((((.((((..............)))).)))).))))))..)))).. ( -32.94) >consensus AAACCACCAACAAGAAU_GCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUUUGAUUCUUUGCAUAACCGAAAUGCAAUUGGGCUUCGACUG .............(((..((((.(((..(((((((((((....))))).(((((......((((.(((......)))...))))...)))))))))))...)))....)))))))..... (-28.59 = -28.65 + 0.06)

| Location | 4,328,796 – 4,328,903 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -30.46 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
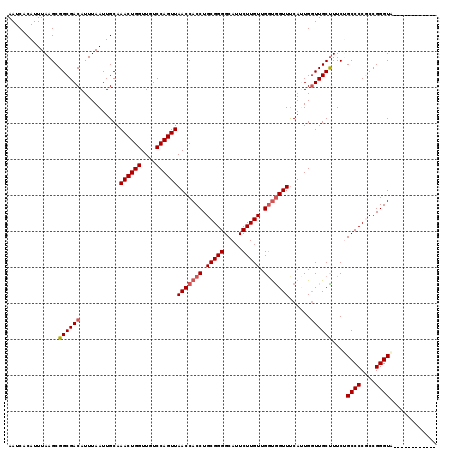
>3L_DroMel_CAF1 4328796 107 + 23771897 AAUCACAUUUAAGCGGCGACAUUUAAUUGCAAACUGGUUGUCCAGUUAACCACCUGCGGGGCAUUCUUGUUGGCAGUUCCAUUGGUUGCUUUCUGCCCCGCCGGGUA------------ .....((.(((((.(....).))))).))..((((((....))))))....((((((((((((.....(..(((((..((...)))))))..)))))))).))))).------------ ( -34.30) >DroSec_CAF1 15367 107 + 1 AAUCACAUUUAAGCGGCGACAUUUAAUUGCAAACUGGUUGUCCAGUUAACCACCUGCGGGGCAUUCUUGUUGGUGGUUUCAUUGGUUGCUUUCAGCCCCGCCGGGUA------------ .............(((((.............((((((....))))))(((((((.(((((.....))))).))))))).....(((((....))))).)))))....------------ ( -35.40) >DroSim_CAF1 13928 107 + 1 AAUCACAUUUAAGCGGCGACAUUUAAUUGCAAACUGGUUGUCCAGUUAACCACCUGCGGGGCAUUCUUGUUGGUGGUUUCAUUGGUUGCUUUCAGCCCCGCCGGGUA------------ .............(((((.............((((((....))))))(((((((.(((((.....))))).))))))).....(((((....))))).)))))....------------ ( -35.40) >DroEre_CAF1 15326 113 + 1 AAUCACAUUUAAGCGGCGACAUUUAAUUGCAAACUGGUUGUCCAGUUAACCACCUGCGGGGCAUUCUUGUUGGUGGUUUCAGUGGUUGCUUUCUGCCCCACCGGGUAUGCACA------ ..........(((((((.((...........((((((....))))))(((((((.(((((.....))))).)))))))...)).)))))))..(((((....)))))......------ ( -38.50) >DroYak_CAF1 14471 119 + 1 AAUCUCAUUUAAGCGGCGACAUUUAAUUGCAAACUGGUUGUCCAGUUAACCACCUGCGGGGCAUUCUUGUUGCUGGUUUCAGUGCUUGCCUUCUGCCCCACCGGGUGUGCACACUCACA ........(((((.(....).))))).((((((((((....))))))...((((((.((((((.....((.((((....))))....))....))))))..))))))))))........ ( -34.30) >consensus AAUCACAUUUAAGCGGCGACAUUUAAUUGCAAACUGGUUGUCCAGUUAACCACCUGCGGGGCAUUCUUGUUGGUGGUUUCAUUGGUUGCUUUCUGCCCCGCCGGGUA____________ ..............((((((...........((((((....))))))(((((((.(((((.....))))).)))))))......))))))....((((....))))............. (-30.46 = -31.10 + 0.64)


| Location | 4,328,796 – 4,328,903 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -32.49 |
| Consensus MFE | -28.17 |
| Energy contribution | -27.97 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
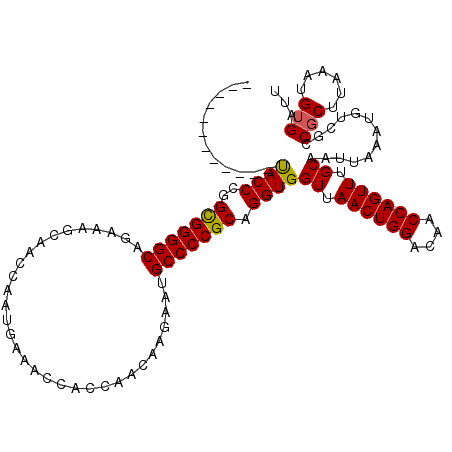
>3L_DroMel_CAF1 4328796 107 - 23771897 ------------UACCCGGCGGGGCAGAAAGCAACCAAUGGAACUGCCAACAAGAAUGCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUU ------------((((..(((((((((........)..(((.....))).......)))))))).))))((.((((((....)))))).)).............(((......)))... ( -32.40) >DroSec_CAF1 15367 107 - 1 ------------UACCCGGCGGGGCUGAAAGCAACCAAUGAAACCACCAACAAGAAUGCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUU ------------((((..(((((((................................))))))).))))((.((((((....)))))).)).............(((......)))... ( -29.05) >DroSim_CAF1 13928 107 - 1 ------------UACCCGGCGGGGCUGAAAGCAACCAAUGAAACCACCAACAAGAAUGCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUU ------------((((..(((((((................................))))))).))))((.((((((....)))))).)).............(((......)))... ( -29.05) >DroEre_CAF1 15326 113 - 1 ------UGUGCAUACCCGGUGGGGCAGAAAGCAACCACUGAAACCACCAACAAGAAUGCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUU ------.(((((((((..((((((((..............................)))))))).))).((.((((((....)))))).))......))).)))(((......)))... ( -30.91) >DroYak_CAF1 14471 119 - 1 UGUGAGUGUGCACACCCGGUGGGGCAGAAGGCAAGCACUGAAACCAGCAACAAGAAUGCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGAGAUU ..((((((.((.((((..((((((((...........(((....))).........)))))))).))))((.((((((....)))))).))...........))))))))......... ( -41.05) >consensus ____________UACCCGGCGGGGCAGAAAGCAACCAAUGAAACCACCAACAAGAAUGCCCCGCAGGUGGUUAACUGGACAACCAGUUUGCAAUUAAAUGUCGCCGCUUAAAUGUGAUU ............((((..(((((((................................))))))).))))((.((((((....)))))).)).............(((......)))... (-28.17 = -27.97 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:00 2006