
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,326,740 – 4,326,934 |
| Length | 194 |
| Max. P | 0.717212 |

| Location | 4,326,740 – 4,326,856 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.11 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4326740 116 - 23771897 AGACUUACGUAUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCACUGGGGGCAAUGUCAAUUUGCGUCGGCCAAGGGAACAGUCAGUCAUCAGUGGACAUCAUCGUCAUCCACUA .((((.(((((..((((((((.(((.(((.(((.....))))))..))).))).)))))....))))).(((...(....).)))))))...((((((...........)))))). ( -37.60) >DroSec_CAF1 13397 112 - 1 AGACAUAUGUAUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCACUGGGGGCAAUGUCAAUUUGCGUCGGCCAAGGGAACAG----UCAUCAGUGGACAUCAUCGUCAUCCACUA .(((..(((((..((((((((.(((.(((.(((.....))))))..))).))).)))))....))))).......(....).)----))...((((((...........)))))). ( -34.30) >DroSim_CAF1 11991 112 - 1 AGACAUGUGUAUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCACUGGGGGCAAUGUCAAUUUGCGUCGGCCAAGGGAACAG----UCAUCAGUGGACAUCAUCGUCAUCCACUA .(((.(((.....((((((((.(((.(((.(((.....))))))..))).))).)))))..((((.(....)))))...))))----))...((((((...........)))))). ( -34.40) >DroEre_CAF1 13258 108 - 1 AGACAU----AUUGACGUGCCGCCACGAGCUGCAAGUUGCACUCACUGGGGGCAAUGUCAAUUUGCCUCGGCCAAGGGAACAG----UCAUCACUGGACACCAUCGUCAUCCAAUA .(((..----(((((((((((.(((.(((.(((.....))))))..))).))).))))))))..((....))...(((..(((----(....))))..).))...)))........ ( -36.10) >DroYak_CAF1 12388 108 - 1 AGACAU----AUUGACGUGCCGCCACGAGCUGCAUGUUGCACUCACUGGGGGCAAUGUCAAUUUGCGUCGGCCAAGGGAACAC----UCAUCAGUGGACAUCAUCGUCAUCCACUA .(((..----(((((((((((.(((.(((.(((.....))))))..))).))).))))))))....))).....(((((.(((----(....))))(((......))).))).)). ( -37.60) >consensus AGACAU__GUAUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCACUGGGGGCAAUGUCAAUUUGCGUCGGCCAAGGGAACAG____UCAUCAGUGGACAUCAUCGUCAUCCACUA .(((.........((((((((.(((.(((.(((.....))))))..))).))).))))).......(((.((.....((........))....)).)))......)))........ (-29.86 = -30.26 + 0.40)


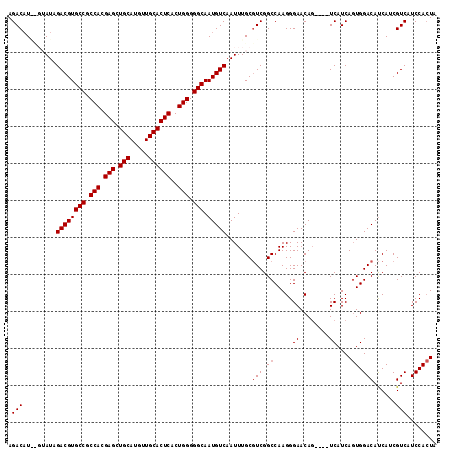
| Location | 4,326,780 – 4,326,896 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -29.14 |
| Energy contribution | -30.75 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4326780 116 - 23771897 CGAACUGAAGGCGGUAAAAGGGGGCGUGGCGGAAACCAAUAGACUUACGUAUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCAC-UGGGGGCAAUGUCAAUUUGCGUCGGCC---AAG ...((((....)))).......(((.(((((.(((.......((....))...((((((((.(((.(((.(((.....))))))..-))).))).)))))..))).))))))))---... ( -36.50) >DroSec_CAF1 13433 116 - 1 CAAAUUGCAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAUAUGUAUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCAC-UGGGGGCAAUGUCAAUUUGCGUCGGCC---AAG ....((((.....)))).....(((.((((....))))...(((.((...((.((((((((.(((.(((.(((.....))))))..-))).))).))))).)).)).))).)))---... ( -40.00) >DroSim_CAF1 12027 116 - 1 CGAAUUGCAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAUGUGUAUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCAC-UGGGGGCAAUGUCAAUUUGCGUCGGCC---AAG ....((((.....)))).....(((.((((....))))...(((..(((.((.((((((((.(((.(((.(((.....))))))..-))).))).))))).)).)))))).)))---... ( -40.50) >DroEre_CAF1 13294 112 - 1 UGCAGGGAAGGCGGUAAAAAGGGGCGUGGCGGAAGCCAAUAGACAU----AUUGACGUGCCGCCACGAGCUGCAAGUUGCACUCAC-UGGGGGCAAUGUCAAUUUGCCUCGGCC---AAG .((.......))(((.....((((((((((....))))........----(((((((((((.(((.(((.(((.....))))))..-))).))).)))))))).)))))).)))---... ( -47.20) >DroYak_CAF1 12424 112 - 1 UGCAUGGAAGGCGGUAAAAAGGGGCGUGGCGGAAGCCAAUAGACAU----AUUGACGUGCCGCCACGAGCUGCAUGUUGCACUCAC-UGGGGGCAAUGUCAAUUUGCGUCGGCC---AAG ....(((..((((.............((((....)))).....((.----(((((((((((.(((.(((.(((.....))))))..-))).))).)))))))).))))))..))---).. ( -45.40) >DroAna_CAF1 13081 101 - 1 ---UCUGGGGGCAUCGAAAAGGGGCGUGGCGGAAGCCAAUAGACAUAC----------------AUUUGUUGCAUGUUGCAGUCACCCGGGGGCAAUGUCGAUUUGAGUCAGCUCUUGAG ---.((((((((.((.....)).((.((((....))))...(((((.(----------------(.....)).))))))).))).)))))((((..((.(.......).))))))..... ( -29.80) >consensus CGAAUUGAAGGCGGUAAAAAGGGGCGUGGCGGAAGCCAAUAGACAUA___AUAGACGUGCCGCCACGAGCUGCAUGUUGCACUCAC_UGGGGGCAAUGUCAAUUUGCGUCGGCC___AAG .........(((.(...(((..((((((((....)))....................((((.(((.(((.(((.....))))))...))).)))))))))..)))....).)))...... (-29.14 = -30.75 + 1.61)



| Location | 4,326,816 – 4,326,934 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.09 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4326816 118 - 23771897 CAUGCUAAACUGAAAAAACA--GAAGAAAAAAAAUUCCAUCGAACUGAAGGCGGUAAAAGGGGGCGUGGCGGAAACCAAUAGACUUACGUAUAGACGUGCCGCCACGAGCUGCAUGUUGC (((((....(((......))--)...........((((.....((((....)))).....))))((((((((.............(((((....)))))))))))))....))))).... ( -29.41) >DroSec_CAF1 13469 116 - 1 CAUGCUAAACUGAAAAAA----ACAGAAAAAAAAGUCCAUCAAAUUGCAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAUAUGUAUAGACGUGCCGCCACGAGCUGCAUGUUGC (((((....(((......----.))).......((..(......((((.....))))..((.(((.((((....))))........((((....))))))).))..)..))))))).... ( -29.70) >DroSim_CAF1 12063 105 - 1 CAUGCUAAACUGAAAA---------------AAAUUCCAUCGAAUUGCAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAUGUGUAUAGACGUGCCGCCACGAGCUGCAUGUUGC (((((...((((....---------------.(((((....))))).....))))....((.(((.((((....))))......((((......))))))).)).......))))).... ( -32.10) >DroEre_CAF1 13330 112 - 1 CAUGCUAAACUAAAAAA----AAACAGAAAAAAAUACCAUUGCAGGGAAGGCGGUAAAAAGGGGCGUGGCGGAAGCCAAUAGACAU----AUUGACGUGCCGCCACGAGCUGCAAGUUGC .................----..................((((((....(((((((..((...((.((((....))))...).)..----.))....))))))).....))))))..... ( -29.20) >DroYak_CAF1 12460 116 - 1 CAUGCUAAACUAAUAAAACGAAAAAAAAAAAAAAUAACAUUGCAUGGAAGGCGGUAAAAAGGGGCGUGGCGGAAGCCAAUAGACAU----AUUGACGUGCCGCCACGAGCUGCAUGUUGC (((((.................................(((((.......))))).....((..((((((((..(.(((((....)----)))).)...))))))))..))))))).... ( -30.90) >consensus CAUGCUAAACUGAAAAAA____AAAAAAAAAAAAUUCCAUCGAAUUGAAGGCGGUAAAAGGGGGCGUGGCGGAAGCCAAUAGACAU__GUAUAGACGUGCCGCCACGAGCUGCAUGUUGC (((((................................(........)..(((((((..........((((....))))...................))))))).......))))).... (-20.69 = -21.09 + 0.40)


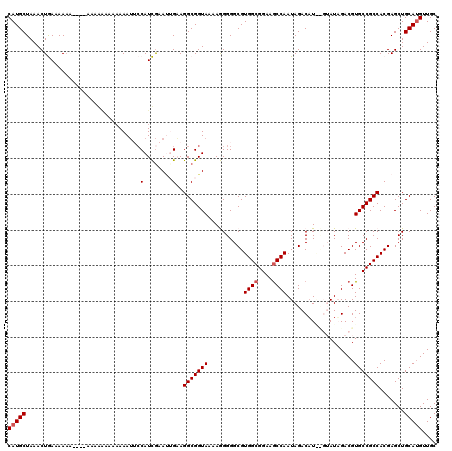
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:54 2006