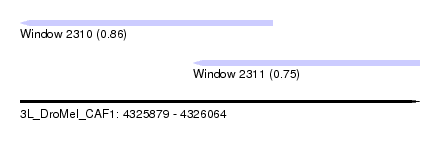
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,325,879 – 4,326,064 |
| Length | 185 |
| Max. P | 0.860451 |

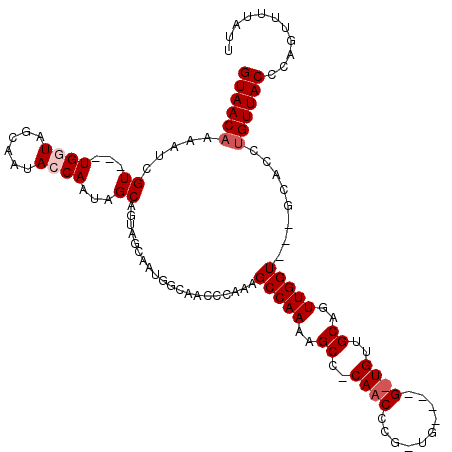
| Location | 4,325,879 – 4,325,996 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -10.94 |
| Energy contribution | -12.24 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4325879 117 - 23771897 GUAACAAAAUCGU---UGGUAGCAAUACCAAUAGCAGUAGCAAUGGCAACCCAAAGCCAAAAGCCUCAACCCGUUGGUUGGUUGUUGCAGUUGGUGCUGCACCUGUUACCCAGUUUUAUU ((((((.....((---(((((....))))))).((((((.((((.(((((....((((((.(((........)))..))))))))))).)))).))))))...))))))........... ( -41.70) >DroSec_CAF1 12560 90 - 1 GUAACAAAAUCGU---UGGUUGCAAUACCAAUAGCAGUAGCAAUGGCAACCCAAAGCCAAAAG------------------------CAGUUGGU---GCACCUGUUACCCAGUUUUAUU ((((((.....((---((((......)))))).(((.((((..((((........))))....------------------------..)))).)---))...))))))........... ( -22.70) >DroSim_CAF1 11192 93 - 1 GUAACAAAAUCGUUGUUGGUUGCAAUACCAAUAGCAGUAGCAAUGGCAACCCAAAGCCAAAAG------------------------CAGUUGGU---GCACCUGUUACCCAGUUUUAUU ((((((.....(((((((((......))))))))).(((.(((((((........))))....------------------------...))).)---))...))))))........... ( -25.21) >DroEre_CAF1 12428 98 - 1 GUAACAAAAUCGU---UGGUAGGAAAAGCAAUAGCAA------------CCCAAAGCCAAAAGCCCCAACCCGCUG----GUUGUUGCAGUUGGU---GCACCUGUUACCCAGUUUUAUU ((((((......(---((((.((....((....))..------------.))...)))))..((.(((((..((..----......)).))))).---))...))))))........... ( -22.50) >DroYak_CAF1 11558 112 - 1 GUAACUAAAUCGUUGUUGGUGGCAACAGCAAUAGCAA-AGCAAAGCAAAGCCAAAGCCAAAAGCCCCAACCCGCUG----GCUGUUGCAGUUGGU---GCACCUGUUACCCAGUUUUAUU (((((......(((((((.((....)).)))))))..-.(((.(((...((...(((((...((........))))----)))...)).)))..)---))....)))))........... ( -30.50) >consensus GUAACAAAAUCGU___UGGUAGCAAUACCAAUAGCAGUAGCAAUGGCAACCCAAAGCCAAAAGCC_CAACCCG_UG____G_UGUUGCAGUUGGU___GCACCUGUUACCCAGUUUUAUU ((((((.....((...((((......))))...))....................(((((..((..((((..........))))..))..)))))........))))))........... (-10.94 = -12.24 + 1.30)

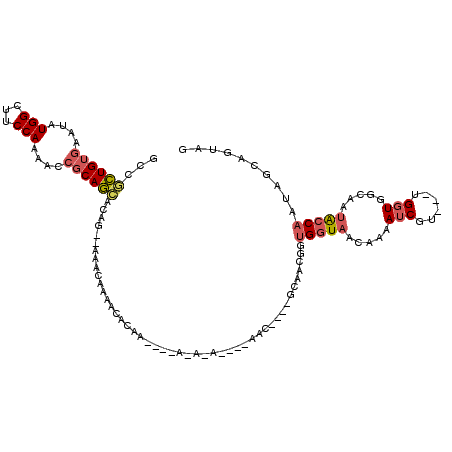
| Location | 4,325,959 – 4,326,064 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.14 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -8.62 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4325959 105 - 23771897 GCCACUGUGAAUAUGGCUUCCAAAACCGCAGUACAG--AAACAAAACACAAA---AAAAA---AAAC----GCAACGGUGGUAACAAAAUCGU---UGGUAGCAAUACCAAUAGCAGUAG ((((((((.....(((...))).....((.......--..............---.....---....----)).)))))))).........((---(((((....)))))))........ ( -20.11) >DroSec_CAF1 12613 97 - 1 GCCGCUGUGAAUAUGGCUUCCAAAACCGCAGCUCAG--AAACAAAACACG--------------AAC----GCGACGGUGGUAACAAAAUCGU---UGGUUGCAAUACCAAUAGCAGUAG ((.(((((...(((.((..((((.((((..((....--............--------------...----))..))))(((......))).)---)))..)).)))...))))).)).. ( -20.31) >DroSim_CAF1 11245 100 - 1 GCCGCUGUGAAUAUGGCUUCCAAAACCGCAGCACAG--AAACAAAACACG--------------AAC----GCGACGGUGGUAACAAAAUCGUUGUUGGUUGCAAUACCAAUAGCAGUAG (((((((((....(((...)))....))))))....--..........((--------------...----.))..)))............(((((((((......)))))))))..... ( -24.70) >DroYak_CAF1 11631 96 - 1 GCCGCUGUGAGUAUGGCUUCCAAAACCGCAGCACUG--AAACAAAACAAAAU---AAAUA------------------UGGUAACUAAAUCGUUGUUGGUGGCAACAGCAAUAGCAA-AG (((((((((.((.(((...)))..))))))))..((--........))....---.....------------------.))).........(((((((.((....)).)))))))..-.. ( -25.00) >DroAna_CAF1 12658 104 - 1 UUCGUUG--CCUAUGGCUUCCAAAACCACAAAACACUAAAAAAAAAGGCUAAUAAACAAAGCCAAACAAAACAAAAAGUGGUAACUAAAUCGU---AGCUGCCACUGCC----------- .......--.....(((.............................((((.........)))).............(((((((.(((.....)---)).))))))))))----------- ( -18.80) >consensus GCCGCUGUGAAUAUGGCUUCCAAAACCGCAGCACAG__AAACAAAACACAA____A_A_A____AAC____GCAACGGUGGUAACAAAAUCGU___UGGUGGCAAUACCAAUAGCAGUAG ...((((((....(((...)))....))))))..............................................(((((.....(((......))).....))))).......... ( -8.62 = -9.30 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:51 2006