
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,324,552 – 4,324,655 |
| Length | 103 |
| Max. P | 0.788082 |

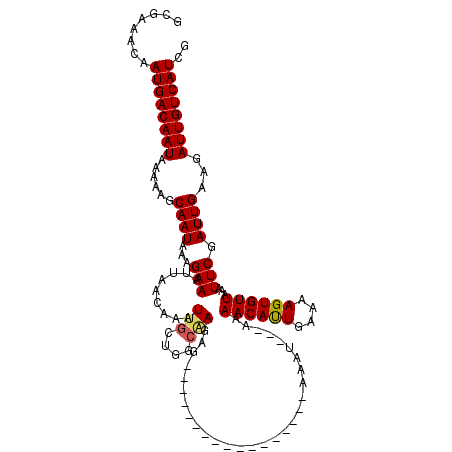
| Location | 4,324,552 – 4,324,655 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.88 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.50 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4324552 103 + 23771897 GCGAAACAAUGACAAUAAAAAGCAAUAAAGAAAUUAACAAAUUGCUGGCAAAAG-----------------AAAAAAAAAAACAUUGAAAAGUGUUAGAUUCGAUUGAAGAUUGUCAUCG ........((((((((......((((...(((.........(((....)))...-----------------.........((((((....))))))...))).))))...)))))))).. ( -17.00) >DroSec_CAF1 11217 100 + 1 GCGAAACAAUGACAAUAAAAAGCAAUAAAGAAAUUAACAAAUUGCUGGCAAGAG-----------------AAAU---AAAACAUUGAAAAGUGUUAGAUUCGAUUGAAGAUUGUCAUCG ........((((((((....((((((..............))))))..(((..(-----------------((..---..((((((....))))))...)))..)))...)))))))).. ( -19.74) >DroSim_CAF1 9826 100 + 1 GCGAAACAAUGACAAUAAAAAGCAAUAAAGAAAUUAACAAAUUACUGGCAAGAG-----------------AAAU---AAAACAUUGAAAAGUGUUAGAUUCGAUUGAAGAUUGUCAUCG ........((((((((......((((...(((.((((((..(((.((.....(.-----------------...)---....)).)))....)))))).))).))))...)))))))).. ( -16.50) >DroEre_CAF1 11091 100 + 1 GCGAAACAAUGACAAUAAAAAGCAAUAAAGAAAUUAACAAAUUGCUGGCGAGGG-----------------GAAU---AAAACAUUAAAAAGUGUUAAAUUCGAUUGAAGAUUGUCAUCG ........((((((((....((((((..............))))))..(((...-----------------((((---..((((((....))))))..))))..)))...)))))))).. ( -17.34) >DroYak_CAF1 10121 100 + 1 GCGAAACAAUGACAAUGAAAAGCAAUAAAGAAAUUAACAAAUUGCUGGCCAGAG-----------------AAAU---GAAACAUUGAAAAGUGUUAAAUUCGAUUGAAGAUUGUCAUCG ........((((((((....((((((..............))))))...(((.(-----------------((..---..((((((....))))))...)))..)))...)))))))).. ( -17.54) >DroAna_CAF1 11477 117 + 1 ACAAAACAAUGACAAUAAAAAGCAAUAAAGAAAUUAACAAAUUGCCGGCAAGAGGCCGAGGAGAGUCGAGAAAAU---AAAACACUGAAAAGUGUUAAAUUCGAUUGAAGAUUGUCAUCG ........((((((((.....(((((..............)))))((((.....)))).....(((((((.....---..((((((....))))))...)))))))....)))))))).. ( -27.14) >consensus GCGAAACAAUGACAAUAAAAAGCAAUAAAGAAAUUAACAAAUUGCUGGCAAGAG_________________AAAU___AAAACAUUGAAAAGUGUUAAAUUCGAUUGAAGAUUGUCAUCG ........((((((((......((((...(((.........(((....))).............................((((((....))))))...))).))))...)))))))).. (-14.16 = -15.50 + 1.34)


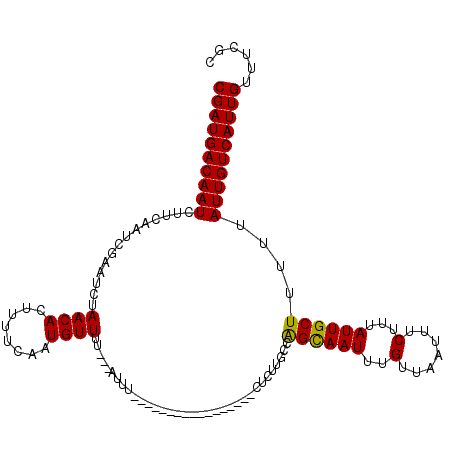
| Location | 4,324,552 – 4,324,655 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.88 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.32 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4324552 103 - 23771897 CGAUGACAAUCUUCAAUCGAAUCUAACACUUUUCAAUGUUUUUUUUUUU-----------------CUUUUGCCAGCAAUUUGUUAAUUUCUUUAUUGCUUUUUAUUGUCAUUGUUUCGC ((((((((((...(((..(((...((((........)))).......))-----------------)..)))..((((((..(.......)...))))))....))))))))))...... ( -14.90) >DroSec_CAF1 11217 100 - 1 CGAUGACAAUCUUCAAUCGAAUCUAACACUUUUCAAUGUUUU---AUUU-----------------CUCUUGCCAGCAAUUUGUUAAUUUCUUUAUUGCUUUUUAUUGUCAUUGUUUCGC ((((((((((...(((..(((...((((........))))..---..))-----------------)..)))..((((((..(.......)...))))))....))))))))))...... ( -15.90) >DroSim_CAF1 9826 100 - 1 CGAUGACAAUCUUCAAUCGAAUCUAACACUUUUCAAUGUUUU---AUUU-----------------CUCUUGCCAGUAAUUUGUUAAUUUCUUUAUUGCUUUUUAUUGUCAUUGUUUCGC ((((((((((...(((..(((...((((........))))..---..))-----------------)..)))..((((((..(.......)...))))))....))))))))))...... ( -13.90) >DroEre_CAF1 11091 100 - 1 CGAUGACAAUCUUCAAUCGAAUUUAACACUUUUUAAUGUUUU---AUUC-----------------CCCUCGCCAGCAAUUUGUUAAUUUCUUUAUUGCUUUUUAUUGUCAUUGUUUCGC ((((((((((........((((..((((........))))..---))))-----------------........((((((..(.......)...))))))....))))))))))...... ( -18.00) >DroYak_CAF1 10121 100 - 1 CGAUGACAAUCUUCAAUCGAAUUUAACACUUUUCAAUGUUUC---AUUU-----------------CUCUGGCCAGCAAUUUGUUAAUUUCUUUAUUGCUUUUCAUUGUCAUUGUUUCGC ((((((((((........(((...((((........))))..---..))-----------------)...((..((((((..(.......)...))))))..))))))))))))...... ( -16.60) >DroAna_CAF1 11477 117 - 1 CGAUGACAAUCUUCAAUCGAAUUUAACACUUUUCAGUGUUUU---AUUUUCUCGACUCUCCUCGGCCUCUUGCCGGCAAUUUGUUAAUUUCUUUAUUGCUUUUUAUUGUCAUUGUUUUGU ((((((((((......((((((..((((((....))))))..---))....)))).......((((.....))))(((((..(.......)...))))).....))))))))))...... ( -25.30) >consensus CGAUGACAAUCUUCAAUCGAAUCUAACACUUUUCAAUGUUUU___AUUU_________________CUCUUGCCAGCAAUUUGUUAAUUUCUUUAUUGCUUUUUAUUGUCAUUGUUUCGC ((((((((((..............((((........))))..................................((((((..(.......)...))))))....))))))))))...... (-13.59 = -13.32 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:49 2006