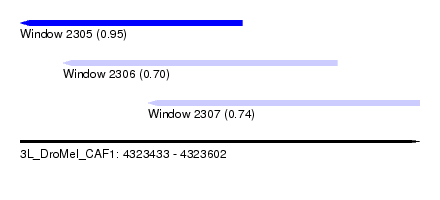
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,323,433 – 4,323,602 |
| Length | 169 |
| Max. P | 0.948616 |

| Location | 4,323,433 – 4,323,527 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -19.90 |
| Energy contribution | -21.10 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4323433 94 - 23771897 CACCCCUGGCCAUAUAUCUAUAUACAUUUAUGUUUCUUUUAUAAUAUAUGUAUAUGUAGGUAGGUAGAGCAGUCGAAGC-----------------AAUUUCCCGCCGGCG .....(((((....((((((((((.((.((((((........)))))).)).))))))))))((.(((((.......))-----------------..))).))))))).. ( -21.00) >DroSec_CAF1 10117 105 - 1 CACCCCUGGCCAUAUAUAUAUAUA------UGUUUUUCUUUUUAUAUAUAUGUAUGUAGGUAGGUAGAGCAGCCGAAGCAGAUCAAUUGGAAAAACAAUUUCCCGCCGGCG ....((((.(((((((((((((((------((..........))))))))))))))).)))))).......((((..((.((..(((((......)))))))..)))))). ( -29.20) >DroSim_CAF1 8738 103 - 1 CACCCCUGGCCAUAUAUAUGUAUA------UGCUUUUCUU--UAUAUAUAUGUAUGUAGGUAGGUAGAGCAGCCGAAGCAGAUCAAUUGGAAAAACAAUUUCCCGCCGGCG ....((((.(((((((((((((((------((........--))))))))))))))).)))))).......((((..((.((..(((((......)))))))..)))))). ( -29.20) >consensus CACCCCUGGCCAUAUAUAUAUAUA______UGUUUUUCUU_UUAUAUAUAUGUAUGUAGGUAGGUAGAGCAGCCGAAGCAGAUCAAUUGGAAAAACAAUUUCCCGCCGGCG .(((((((..((((((((((((((..................))))))))))))))))))..)))......((((..((.....(((((......)))))....)))))). (-19.90 = -21.10 + 1.20)


| Location | 4,323,451 – 4,323,567 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -17.49 |
| Energy contribution | -19.93 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4323451 116 - 23771897 AUUCCAUUUAAUCAAACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUCUAUAUACAUUUAUGUUUCUUUUAUAAUAUAUGUAUAUGUAGGUAGGUAGAGCAGUCGA---- .((((((((.........)))))))).(((((((....)))....(((.((...((((((((((.((.((((((........)))))).)).)))))))))))))))..))))...---- ( -25.40) >DroSec_CAF1 10152 110 - 1 AUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUAUAUAUA------UGUUUUUCUUUUUAUAUAUAUGUAUGUAGGUAGGUAGAGCAGCCGA---- .((((((((.........))))))))...((((.....(((.(..((.((((((((((((((((------((..........))))))))))))))).))).))..).))))))).---- ( -31.10) >DroSim_CAF1 8773 108 - 1 AUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUAUGUAUA------UGCUUUUCUU--UAUAUAUAUGUAUGUAGGUAGGUAGAGCAGCCGA---- .((((((((.........))))))))...((((..((.(((...((((..((((((((((((((------..........--))))))))))))))))))...)))..)).)))).---- ( -31.10) >DroEre_CAF1 10016 105 - 1 AUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCUCCCCUGGCCAUGUA----UA--------UGUAUUUCUU--UAUAUAUGU-UAUGUAGGUAGGUAGAGCAGUCGAUUGG .((((((((.........))))))))........((..(((((..((.((((((((----((--------(((((.....--.))))))).-))))).))).))..)))))..))..... ( -26.90) >DroYak_CAF1 9028 95 - 1 AUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUACACCCCUGGCCAUAAAUAUGUA--------UGUAUUUCUU--UAUAUAU----AUGUAGGUAGGUAGAG----------- .((((((((.........))))))))..................((((.((....(((((((--------((((......--)))))))----)))).)))))).....----------- ( -19.40) >consensus AUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUAUAUAUA______UGUUUUUCUU__UAUAUAUAU_UAUGUAGGUAGGUAGAGCAGCCGA____ .((((((((.........))))))))....(((..((.(((...((((..(((((((((((((....................)))))))))))))))))...)))..)).)))...... (-17.49 = -19.93 + 2.44)



| Location | 4,323,487 – 4,323,602 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4323487 115 - 23771897 ACUUUCCUGACCCA-ACGAGAGGCGGGCAAUUUGCCAUUCCAUUUAAUCAAACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUCUAUAUACAUUUAUGUUUCUUUU ..............-..((((((((((((...(((((((((((((.........))))))))...)))))....))).)).......((((....))))........))))))).. ( -23.10) >DroSec_CAF1 10188 109 - 1 ACUUUCCUGCCCCA-ACAAGCGGCGGGCAAUUUGCCAUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUAUAUAUA------UGUUUUUCUU ......(((((.(.-....).)))))(((...(((((((((((((.........))))))))...)))))....))).........((((((....))))------))........ ( -25.30) >DroSim_CAF1 8807 109 - 1 ACUUUCCUGCCCCA-ACGAGCGGCGGGCAAUUUGCCAUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUAUGUAUA------UGCUUUUCUU ......(((((.(.-....).)))))(((...(((((((((((((.........))))))))...)))))....)))......(((.(((((....))))------))))...... ( -26.20) >DroEre_CAF1 10053 104 - 1 ACUUUCCUGCCCCAAACAAGCGGCAGGCGAUUUGCCAUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCUCCCCUGGCCAUGUA----UA--------UGUAUUUCUU .....((((((.(......).)))))).....(((((((((((((.........))))))))...))))).(((((((.....)).))))).----..--------.......... ( -27.40) >DroYak_CAF1 9051 108 - 1 ACUUUCCUGCCCCAAACAAGCGGCGGGCAAUUUGCCAUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUACACCCCUGGCCAUAAAUAUGUA--------UGUAUUUCUU .....((((((.(......).)))))).....(((((((((((((.........))))))))...))))).(((((((................))))--------)))....... ( -27.19) >consensus ACUUUCCUGCCCCA_ACAAGCGGCGGGCAAUUUGCCAUUCCAUUUAAUCAGACAAGAUGGAAAACUGGCACGCAUGCACCCCUGGCCAUAUAUAUAUAUA______UGUUUUUCUU .....((((((.(......).)))))).....(((((((((((((.........))))))))...))))).............................................. (-21.10 = -21.14 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:48 2006