| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,310,641 – 4,310,749 |
| Length | 108 |
| Max. P | 0.713712 |

| Location | 4,310,641 – 4,310,749 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
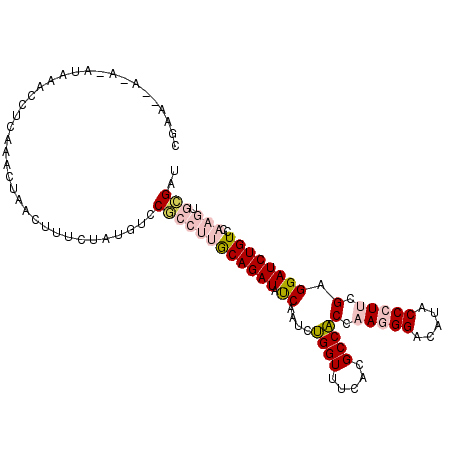
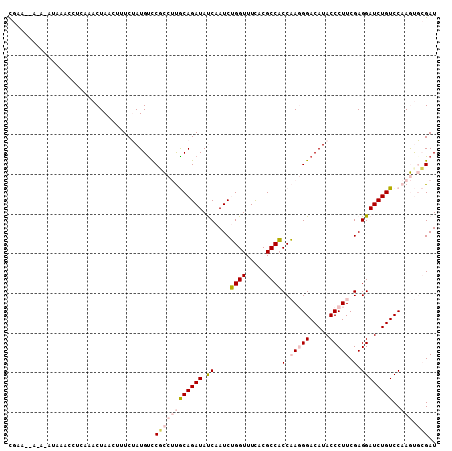
>3L_DroMel_CAF1 4310641 108 - 23771897 CGAAUAAUACAUAAACCUCAAACUAACUCUCUAUGUCCGCCUUGCAGAUAUCAAUCUGGUUUCACGCCACCAAGGGACAUACCCUUCGAGGAUCUGUCCAAGUGCGAU .....................................((((((((((((.((....((((.....))))..(((((.....))))).))..)))))..)))).))).. ( -22.10) >DroPse_CAF1 5141 88 - 1 CCAU----------------AUCUAA---UCAAUGUCCUUC-CACAGAUACCAAUCGGGUUUCGCGCCCCCCACGGACAUACCCUUCGAGGAUCUGUCCAAACUAGAU ....----------------(((((.---...((((((...-....(((....)))((((.....)))).....)))))).........(((....)))....))))) ( -18.50) >DroSec_CAF1 4856 108 - 1 UGAAAUAUUAAUAAACCCCAAACUAACUUUCUAUGUCCGCCUUGCAGAUAUCAAUCUGGUUUCACGCCACCAAGGGACAUACCCUUCGAGGAUCUGUCCAAGUGCGAU .....................................((((((((((((.((....((((.....))))..(((((.....))))).))..)))))..)))).))).. ( -22.10) >DroSim_CAF1 3561 108 - 1 CGAAGAACACAUAAACCUCAAACUAACUUUCUAUGUUCGCCUUGCAGAUAUCAAUCUGGUUUCACGCCACCAAGGGACAUACCCUUCGAGGAUCUGUCCAAGUGCGAU ....................................(((((((((((((.((....((((.....))))..(((((.....))))).))..)))))..)))).)))). ( -23.40) >DroYak_CAF1 3498 108 - 1 CGAAAGUAAAAUGGAUCUUAAACUAACAUUCUAUGCCCACCUUGCAGAUAUCAAUCUGGUUUUACGCCACCAAGGGACAUACCCUUCGAGGAUCUGUCCAAGUGCGAU .....(((...(((((..........(((...)))....((((((((((....))))).............(((((.....))))))))))....)))))..)))... ( -19.80) >DroPer_CAF1 19062 88 - 1 CCAU----------------AUCUAA---UCAAUGUUCUUC-CACAGAUACCAAUCGGGUUUCGCGCCCCCCACGGACAUACCCUUCGAGGAUCUGUCCAAACUAGAU ....----------------(((((.---............-..............((((.....)))).....(((((...((.....))...)))))....))))) ( -15.80) >consensus CGAA__A_A_AUAAACCUCAAACUAACUUUCUAUGUCCGCCUUGCAGAUAUCAAUCUGGUUUCACGCCACCAAGGGACAUACCCUUCGAGGAUCUGUCCAAGUGCGAU .....................................((((((((((((.((....((((.....))))(.(((((.....))))).).))))))))..))).))).. (-14.62 = -15.57 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:44 2006