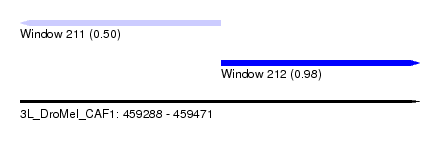
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 459,288 – 459,471 |
| Length | 183 |
| Max. P | 0.977018 |

| Location | 459,288 – 459,380 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -16.82 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 459288 92 - 23771897 CGGGUGCGGUUCAGGCGCAACUUUAAUUUAUGAGCUGCUC--AAAAAAA--AAAAAAAAUUUACGA------UAAUUUGUAGUUGCAGCCGAGGAUUGUCGG ((...(((((((.((((((((.....(((.((((...)))--).)))..--..........(((((------....)))))))))).)))..))))))))). ( -22.10) >DroSec_CAF1 40301 95 - 1 CGGGUGCGGUUCAGGCGCAACUUUAAUUUAUGAGCUGCUC--AAAAAAA-----AAAAGUGUACAAUACUCGUAAUUUGUAGUUGCAGCCGAGGAUUGCCGG ...(.(((((((.(((((((((.(((.(((((((.((..(--(......-----.....))..))...))))))).))).)))))).)))..)))))))).. ( -28.50) >DroSim_CAF1 41200 93 - 1 CGGGUGCGGUUCAGGCGCAACUUUAAUUUAUGAGCUGCUC--AAAAA-------AAAAGUGUACAAUACUCAUAAUUUGUAGUUGCAGCCGAGGAUUGCCGA ...(.(((((((.(((((((((.(((.(((((((.((..(--(....-------.....))..))...))))))).))).)))))).)))..)))))))).. ( -29.10) >DroEre_CAF1 42222 94 - 1 CGGGUGCGGUUCAGGCGCAACUUUAAUUUAUGAGUUGCUC--AAAAAAUACAAAAAAAAAGCACAA------UAAUUUGUAGUUGCAGCCGAGGAUUGGCGG ...((.((((((.(((((((((.(((.((((....((((.--.................))))..)------))).))).)))))).)))..)))))))).. ( -22.67) >DroYak_CAF1 41734 96 - 1 CGGGUGCGGUUCGGGCGCAACUUUAAUUUAUGAGUUGCUCACAAAAAAAACAGAAAAUGUGCACAA------UAAUUUGUAGCUGCAGCCGAGGAUUGUCGG (((.(((((((...(((((((((........)))))))...........(((.....)))))((((------....))))))))))).)))........... ( -27.50) >consensus CGGGUGCGGUUCAGGCGCAACUUUAAUUUAUGAGCUGCUC__AAAAAAA__A_AAAAAGUGUACAA______UAAUUUGUAGUUGCAGCCGAGGAUUGCCGG ((...(((((((.(((((((((.........(((...)))......................((((..........)))))))))).)))..))))))))). (-16.82 = -16.90 + 0.08)

| Location | 459,380 – 459,471 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 459380 91 + 23771897 UCCAUCAAUUUUUAUUUGCGCCUUCAACUUGAGAUGUUCAAUUUUUCGGGGCUUGUCUGCCGGAAAACUGAUAUUGUUGCGCAGAUAAUGC ...........((((((((((..((.....))(((((.((.((((((((.(......).)))))))).)))))))...))))))))))... ( -23.40) >DroSec_CAF1 40396 91 + 1 UCCAUCAAUUUUUAUUUGCGCCUUCAACUUGAGAUGUUCAAUUUUUCGGGGCUUGUCUGCCGGAAAACUGAUAUUGUUGCGCAGAUAAUUC ...........((((((((((..((.....))(((((.((.((((((((.(......).)))))))).)))))))...))))))))))... ( -23.40) >DroSim_CAF1 41293 91 + 1 UCCAUCAAUUUUUAUUUGCGCCUUCAACUUGAGAUGUUCAAUUUUUCGGGGCUUGUCUGCCGGAAAACUGAUAUUGUUGCGCAGAUAAUUC ...........((((((((((..((.....))(((((.((.((((((((.(......).)))))))).)))))))...))))))))))... ( -23.40) >DroEre_CAF1 42316 91 + 1 UCCAUCAAAUUUUAUUUGCGCCUUCAACUUGAGAUGCACAAUUUUUCGGGGCUUGUCUGCCGGAAAACUGAUAUUGUUGUGCAGAUAAUGU ...........((((((((((.(((.....))).....((.((((((((.(......).)))))))).))........))))))))))... ( -19.70) >DroYak_CAF1 41830 91 + 1 UCCAUCAACUUUUAUUUGCGCCUUCAACUUGAGAUGUACAACUUUUCGGGGCUUGUCUGCCGGAAAACUGAUAUUGUUGUGCAGAUAAUGU ...........((((((((((..((.....))(((((.((..(((((((.(......).)))))))..)))))))...))))))))))... ( -19.80) >consensus UCCAUCAAUUUUUAUUUGCGCCUUCAACUUGAGAUGUUCAAUUUUUCGGGGCUUGUCUGCCGGAAAACUGAUAUUGUUGCGCAGAUAAUGC ...........((((((((((..((.....))(((((.((.((((((((.(......).)))))))).)))))))...))))))))))... (-21.54 = -21.70 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:51 2006