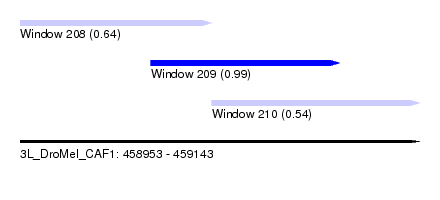
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 458,953 – 459,143 |
| Length | 190 |
| Max. P | 0.993992 |

| Location | 458,953 – 459,044 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -12.84 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
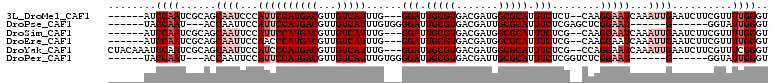
>3L_DroMel_CAF1 458953 91 + 23771897 AUCUGCCCACAGAUGCGAGUACAACAUGCAACAUUCGAGGACGGUAUGGGAUGGGAUGGC---CACCGAACCAUCUUAGACC-----AUCCCAUUCCAA------ .((((....))))((((.(.....).))))............((.((((((((((((((.---.......))))).....))-----))))))).))..------ ( -28.50) >DroSec_CAF1 39977 81 + 1 AUCUGCCCACAGAUGCGAGUACAACAUGCAACAUUCGAGG----------AUGGGAUGGC---CACCGAACCAUACUAUACC-----AUCCCAUUCGAA------ .((((....))))((((.(.....).))))........((----------(((((((((.---.................))-----)))))))))...------ ( -22.57) >DroSim_CAF1 40876 81 + 1 AUCUGCCCACAGAUGCGAGUACAACAUGCAACAUUCGAGG----------AUGGGAUGGC---CACCGAACCAUACUAGACC-----AUCCCAUUCGAA------ .((((....))))((((.(.....).))))........((----------(((((((((.---(..............).))-----)))))))))...------ ( -22.84) >DroYak_CAF1 41379 95 + 1 AUCUGCCCACAGAUGCGAGUACAACAUGCAACAUCUGAGG----------AUGGGAUGGUGGCCAUCGAACCAUCCUAGACCAUCCCAUCCCAUUCCAAAUCCAA .........((((((...(((.....)))..)))))).((----------((((((((((((........)).......)))))))))))).............. ( -28.41) >consensus AUCUGCCCACAGAUGCGAGUACAACAUGCAACAUUCGAGG__________AUGGGAUGGC___CACCGAACCAUACUAGACC_____AUCCCAUUCCAA______ .((((....))))((((.(.....).))))......(.((...........((((((((...........))))))))...........)))............. (-12.84 = -13.15 + 0.31)

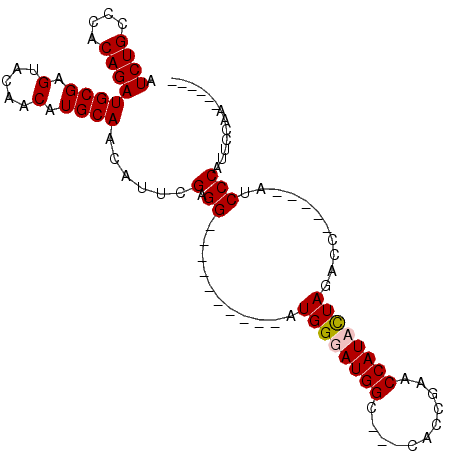
| Location | 459,015 – 459,105 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.24 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 459015 90 + 23771897 CCGAACCAUCUUAGACC-----AUCCCAUUCCAA------------AUCCAAUCGCAGCAAUCCCAUUCCAUGACGUUGUCAUUUGGGAUUGCGUGACGAUGGCGCA ..(..(((((...(...-----..).........------------......((((.(((((((((....(((((...))))).))))))))))))).)))))..). ( -27.30) >DroSec_CAF1 40029 90 + 1 CCGAACCAUACUAUACC-----AUCCCAUUCGAA------------AUCCAAUCGCAGCAAUUCCAUUCCAUGACGUUGUCAUUUGGGAUUGCGUGACGAUGGCGCA ...............((-----(((......(..------------...)..((((.(((((((((....(((((...))))).))))))))))))).))))).... ( -23.20) >DroSim_CAF1 40928 90 + 1 CCGAACCAUACUAGACC-----AUCCCAUUCGAA------------AUCCAAUCGCAGCAAUUCCAUUCCAUGACGUUGUCAUUUGGGAUUGCGUGACGAUGGCGCA .............(.((-----(((......(..------------...)..((((.(((((((((....(((((...))))).))))))))))))).))))))... ( -23.80) >DroEre_CAF1 41964 75 + 1 --------------------CCAUCCCAUUCCCA------------AUCCAAUCGCAGCAAUUCCAACCCAUGACGUUGUCAUUUGGGAUUGCGUGACGAUGGCGCA --------------------..............------------..(((.(((((((((((((((...(((((...))))))))))))))).)).)))))).... ( -22.50) >DroYak_CAF1 41434 107 + 1 UCGAACCAUCCUAGACCAUCCCAUCCCAUUCCAAAUCCAACUACAAAUCCAAUCGCAGCAAUUCCAUCCCAUGACGUUGUCAUUUGGGAUUGCGUGACGAUGGCGCA ..(..(((((...((........))...........................((((.(((((((((....(((((...))))).))))))))))))).)))))..). ( -24.40) >consensus CCGAACCAUACUAGACC_____AUCCCAUUCCAA____________AUCCAAUCGCAGCAAUUCCAUUCCAUGACGUUGUCAUUUGGGAUUGCGUGACGAUGGCGCA ................................................(((.((((((((((((((....(((((...))))).))))))))).)).)))))).... (-22.40 = -22.24 + -0.16)

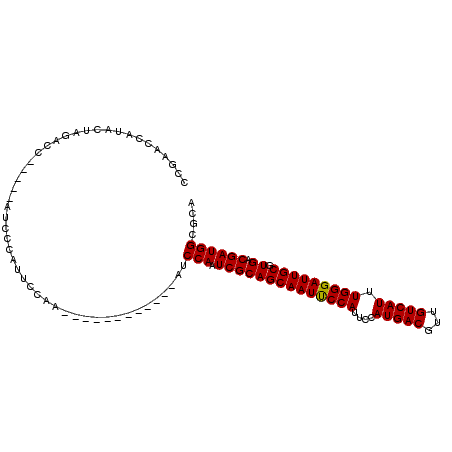
| Location | 459,044 – 459,143 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.81 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
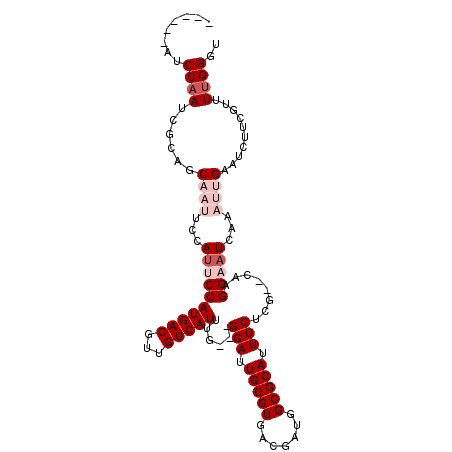
>3L_DroMel_CAF1 459044 99 + 23771897 ------AUCCAAUCGCAGCAAUCCCAUUCCAUGACGUUGUCAUUUG---GGAUUGCGUGACGAUGGCGCAUUUCUCU--CAAGGAAUCAAAUUGAAUCUUCGUUUUGGGU ------((((((((((.(((((((((....(((((...))))).))---)))))))))))((((...(.((((((..--..)))))))..))))..........)))))) ( -29.90) >DroPse_CAF1 101536 89 + 1 ------UACCAAU---ACCAAUUCCAUUCCAUGACGUUGUCAUUUGUGGGGAUUGCGUGACGAUUGCGCAUUUCUCGAGCUCGGAAU------G------GGUAUUGGGU ------..(((((---(((.(((((.....(((((...)))))...((((((.((((..(...)..)))).)))))).....)))))------.------)))))))).. ( -34.70) >DroSim_CAF1 40957 99 + 1 ------AUCCAAUCGCAGCAAUUCCAUUCCAUGACGUUGUCAUUUG---GGAUUGCGUGACGAUGGCGCAUUUCUCG--CAAGGAAUCAAAUUGAAUCUUCGUUUUGGGU ------((((((((((.(((((((((....(((((...))))).))---)))))))))))(((.((..((.(((.(.--...))))......))..)).)))..)))))) ( -29.00) >DroEre_CAF1 41978 99 + 1 ------AUCCAAUCGCAGCAAUUCCAACCCAUGACGUUGUCAUUUG---GGAUUGCGUGACGAUGGCGCAUUUCUCG--CAAGGAAUCAAAUUGAAUCUUCGUUUUGGGU ------((((((((((.((((((((((...(((((...))))))))---)))))))))))(((.((..((.(((.(.--...))))......))..)).)))..)))))) ( -29.70) >DroYak_CAF1 41474 105 + 1 CUACAAAUCCAAUCGCAGCAAUUCCAUCCCAUGACGUUGUCAUUUG---GGAUUGCGUGACGAUGGCGCAUUUCUCG--CCAGGAAUCAAAUUGAAUCUUCGUUUCGGGU ......((((.......(((((((((....(((((...))))).))---)))))))(.((((((((((.......))--)))(((.((.....)).)))))))).))))) ( -31.70) >DroPer_CAF1 98746 89 + 1 ------UACCAAU---ACCAAUUCCAUUCCAUGACGUUGUCAUUUGUGGGGAUUGCGUGACGAUUGCGCAUUUCUCGGUCUCGGAAU------G------GGUAUUGGGU ------..(((((---(((.(((((...(((((((...)))))....(((((.((((..(...)..)))).)))))))....)))))------.------)))))))).. ( -35.80) >consensus ______AUCCAAUCGCAGCAAUUCCAUUCCAUGACGUUGUCAUUUG___GGAUUGCGUGACGAUGGCGCAUUUCUCG__CAAGGAAUCAAAUUGAAUCUUCGUUUUGGGU ........((((......((((...((((((((((...)))))......(((.(((((.......))))).)))........)))))...))))..........)))).. (-14.14 = -15.81 + 1.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:49 2006