
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,264,317 – 4,264,413 |
| Length | 96 |
| Max. P | 0.959259 |

| Location | 4,264,317 – 4,264,413 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -17.16 |
| Energy contribution | -16.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
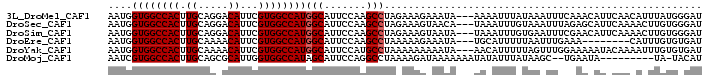
>3L_DroMel_CAF1 4264317 96 + 23771897 AUCCCAUAAAUGUUGAAUGUUUGAAAUUUAUAAAUUUU---UAUUUCUUUCUAGGCUUGGAAUGCCAUGGCCACGAAUGUCCUGCAAGUGGCCACCAUU .....((((((.(..(....)..).)))))).......---............(((.......))).(((((((...((.....)).)))))))..... ( -19.50) >DroSec_CAF1 28665 96 + 1 AUCCCACAAGUUUUGAAUGCUCUAAAUUUACAAAUUUA---UGUUACUUUCUAGGCUUGGAAUGCCAUGGCCACGAAUGUCCUGCAAGUGGCCACCAUU ......(((((((.(((.((..((((((....))))))---.))....))).))))))).((((...(((((((...((.....)).))))))).)))) ( -22.60) >DroSim_CAF1 28526 96 + 1 AUCCCACAAGUUUUGAAUGUUCGAAAUUCACAAAUUUA---UAUUACUUUCUAGGCUUGGAAUGCCAUGGCCACGAAUGUCCUGCAAGUGGCCACCAUU ......(((((((.(((.((...(((((....))))).---....)).))).))))))).((((...(((((((...((.....)).))))))).)))) ( -22.70) >DroEre_CAF1 29409 88 + 1 AUCACACAAAUG--------UUUCAAAUUAAAAAUGCA---UAUUUCUUUUUAGGCUUGGAAUGCCAUGGCCACGAAUGUUUUGCAAGUGGCCACCAUU ...........(--------((((((.(((((((....---......)))))))..)))))))....(((((((...((.....)).)))))))..... ( -18.80) >DroYak_CAF1 31273 96 + 1 AUCACACAAAUUUUGUAUUUUUCCAAACUAAAAAUGUU---UAUUUUUUUUUAGGCAUGGAAUGCCAUGGCCACGAAUGUUUUGCAAGUGGCCACCAUU ....................(((((..(((((((....---......)))))))...))))).....(((((((...((.....)).)))))))..... ( -21.80) >DroMoj_CAF1 25861 87 + 1 AUGUA-UA---------UAUUCA--GCUUAUAAAUAUAUUUUUUUAUCUUUUAGGCCUGGAAUGCUAUGGCCACCAAUGCGCUGCAAGUGGCCACGAUU ..(((-(.---------..((((--(((((.(((.(((......))).))))))).)))))))))..(((((((...(((...))).)))))))..... ( -20.60) >consensus AUCCCACAAAUUUUGAAUGUUCGAAAUUUAAAAAUUUA___UAUUACUUUCUAGGCUUGGAAUGCCAUGGCCACGAAUGUCCUGCAAGUGGCCACCAUU .....................................................(((.......))).(((((((...(((...))).)))))))..... (-17.16 = -16.88 + -0.28)



| Location | 4,264,317 – 4,264,413 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -16.03 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
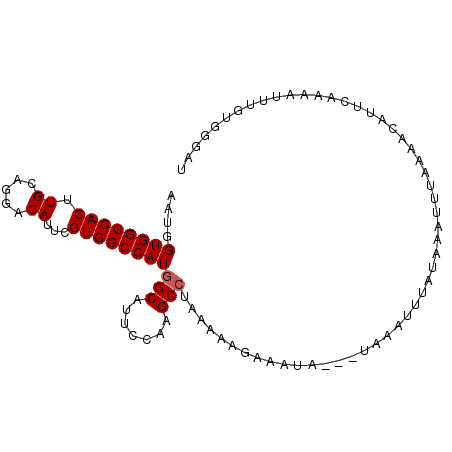
>3L_DroMel_CAF1 4264317 96 - 23771897 AAUGGUGGCCACUUGCAGGACAUUCGUGGCCAUGGCAUUCCAAGCCUAGAAAGAAAUA---AAAAUUUAUAAAUUUCAAACAUUCAACAUUUAUGGGAU ((((((((((((.((.....))...))))))))(((.......)))..((((...(((---(....))))...))))...))))............... ( -19.10) >DroSec_CAF1 28665 96 - 1 AAUGGUGGCCACUUGCAGGACAUUCGUGGCCAUGGCAUUCCAAGCCUAGAAAGUAACA---UAAAUUUGUAAAUUUAGAGCAUUCAAAACUUGUGGGAU ..(.((((((((.((.....))...)))))))).).((((((.(((((((...(.(((---......))).).))))).))...((.....)))))))) ( -21.30) >DroSim_CAF1 28526 96 - 1 AAUGGUGGCCACUUGCAGGACAUUCGUGGCCAUGGCAUUCCAAGCCUAGAAAGUAAUA---UAAAUUUGUGAAUUUCGAACAUUCAAAACUUGUGGGAU ..(.((((((((.((.....))...)))))))).).((((((......((((...(((---......)))...))))((....))........)))))) ( -20.50) >DroEre_CAF1 29409 88 - 1 AAUGGUGGCCACUUGCAAAACAUUCGUGGCCAUGGCAUUCCAAGCCUAAAAAGAAAUA---UGCAUUUUUAAUUUGAAA--------CAUUUGUGUGAU ....((((((((.((.....))...))))))))(((.......)))..........((---((((.((((.....))))--------....)))))).. ( -18.10) >DroYak_CAF1 31273 96 - 1 AAUGGUGGCCACUUGCAAAACAUUCGUGGCCAUGGCAUUCCAUGCCUAAAAAAAAAUA---AACAUUUUUAGUUUGGAAAAAUACAAAAUUUGUGUGAU ..(.((((((((.((.....))...)))))))).)..(((((...(((((((......---....)))))))..)))))..(((((.....)))))... ( -23.10) >DroMoj_CAF1 25861 87 - 1 AAUCGUGGCCACUUGCAGCGCAUUGGUGGCCAUAGCAUUCCAGGCCUAAAAGAUAAAAAAAUAUAUUUAUAAGC--UGAAUA---------UA-UACAU .(((((((((((((((...)))..))))))))).((.......))......)))......(((((((((.....--))))))---------))-).... ( -21.90) >consensus AAUGGUGGCCACUUGCAGGACAUUCGUGGCCAUGGCAUUCCAAGCCUAAAAAGAAAUA___UAAAUUUAUAAAUUUAAAACAUUCAAAAUUUGUGGGAU ....((((((((.((.....))...))))))))(((.......)))..................................................... (-16.03 = -16.20 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:30 2006