
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,256,181 – 4,256,409 |
| Length | 228 |
| Max. P | 0.996951 |

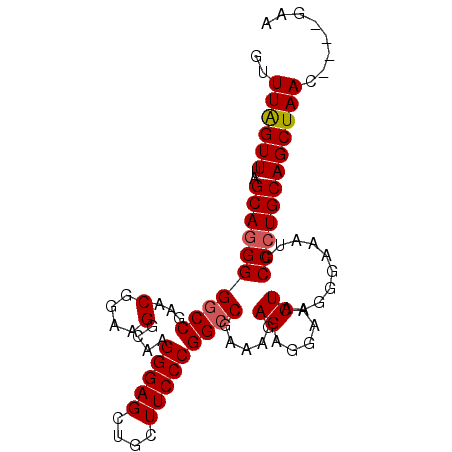
| Location | 4,256,181 – 4,256,276 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.90 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4256181 95 + 23771897 GUUUAGUUUAGCAGGGGGCCAAACGGAAGGCAAGGGAGCUGCUUCCCGGCCGAAAGAGAGGAACUAAGGGAAAUGCCCUGCAGCUAACUGCUAAA ......((((((((...(((........))).....(((((((((((...(....).).))))...((((.....))))))))))..)))))))) ( -33.40) >DroSec_CAF1 20568 91 + 1 GUUUAGUUUAGCAGGGGGCCGAACGGAAGGCAAGGGAGCUGCUUCCCGGCCGAAAGAGAGGAACUAAGGGAAAUGCCCUGCAGCUAAC----GAA ..((((((..(((((((((((...(((((.((.(....))))))))))))).....((.....))..........)))))))))))).----... ( -30.50) >DroSim_CAF1 20390 91 + 1 GUUUAGUUUAGCACGGGGCCGAACGGAAGGCAAGGGAGCUGCUUCCCGGCCGAAAGAGAGGAACUAAGGGAAAUGCCCUGCAGCUAAC----GAA ((((.(((..((.....))..)))...))))..(..(((((((((((...(....).).))))...((((.....))))))))))..)----... ( -27.80) >DroEre_CAF1 21262 91 + 1 GUUUAGUUUAGCAGGGGACCAAACGGAAGGCAAGGGAGCUGCUUCCCGGCCGAAAGAGAGGAACUAAGGGAAAUGCCCUGCAGCUAAC----GAA ..((((((..((((((.(((...(((.(((((.(....)))))).))).((........)).......))...).)))))))))))).----... ( -27.60) >DroYak_CAF1 23088 91 + 1 GUUUGGUUUAGCAGGGGGCCGAACGGAAGGCAAGGGAGCUGCUUCCCGGCCGAAAGAGAGGAACUAAGGGAAAUGCCCUGCAGCUAAC----GAA (((((((((......))))))))).........(..(((((((((((...(....).).))))...((((.....))))))))))..)----... ( -31.10) >consensus GUUUAGUUUAGCAGGGGGCCGAACGGAAGGCAAGGGAGCUGCUUCCCGGCCGAAAGAGAGGAACUAAGGGAAAUGCCCUGCAGCUAAC____GAA ..((((((..((((((((((...(....)....(((((....))))))))).....((.....))..........))))))))))))........ (-27.48 = -27.72 + 0.24)

| Location | 4,256,181 – 4,256,276 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 95.90 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -23.90 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4256181 95 - 23771897 UUUAGCAGUUAGCUGCAGGGCAUUUCCCUUAGUUCCUCUCUUUCGGCCGGGAAGCAGCUCCCUUGCCUUCCGUUUGGCCCCCUGCUAAACUAAAC ((((((((..(((((.((((.....)))))))))..........((((((((((..((......)))))))...)))))..))))))))...... ( -30.90) >DroSec_CAF1 20568 91 - 1 UUC----GUUAGCUGCAGGGCAUUUCCCUUAGUUCCUCUCUUUCGGCCGGGAAGCAGCUCCCUUGCCUUCCGUUCGGCCCCCUGCUAAACUAAAC ...----.((((..((((((..........((.....)).....((((((((((..((......)))))))...)))))))))))....)))).. ( -26.80) >DroSim_CAF1 20390 91 - 1 UUC----GUUAGCUGCAGGGCAUUUCCCUUAGUUCCUCUCUUUCGGCCGGGAAGCAGCUCCCUUGCCUUCCGUUCGGCCCCGUGCUAAACUAAAC ...----.(((((.((.((((.........((.....))....((..(((((.((((.....)))).)))))..)))))).)))))))....... ( -26.20) >DroEre_CAF1 21262 91 - 1 UUC----GUUAGCUGCAGGGCAUUUCCCUUAGUUCCUCUCUUUCGGCCGGGAAGCAGCUCCCUUGCCUUCCGUUUGGUCCCCUGCUAAACUAAAC ...----(..((((((((((.....))))...(((((..(.....)..)))))))))))..).........(((((((.....)))))))..... ( -23.80) >DroYak_CAF1 23088 91 - 1 UUC----GUUAGCUGCAGGGCAUUUCCCUUAGUUCCUCUCUUUCGGCCGGGAAGCAGCUCCCUUGCCUUCCGUUCGGCCCCCUGCUAAACCAAAC ...----.......((((((..........((.....)).....((((((((((..((......)))))))...))))))))))).......... ( -25.50) >consensus UUC____GUUAGCUGCAGGGCAUUUCCCUUAGUUCCUCUCUUUCGGCCGGGAAGCAGCUCCCUUGCCUUCCGUUCGGCCCCCUGCUAAACUAAAC ........((((..((((((..........((.....)).....((((((((((..((......)))))))...)))))))))))....)))).. (-23.90 = -23.90 + 0.00)


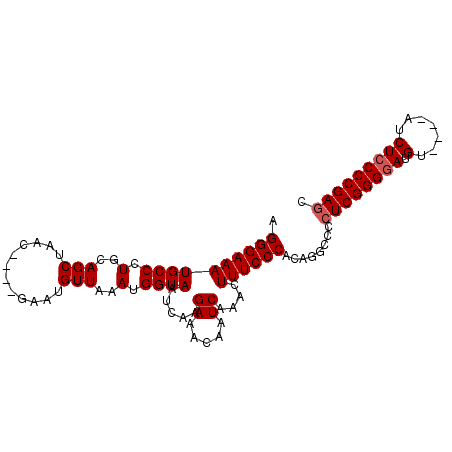
| Location | 4,256,247 – 4,256,343 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4256247 96 + 23771897 AGGGAAAUGCCCUGCAGCUAACUGCUAAAUGUUAAAUGGUAAUCAAGAAACAAUCAAACUUUCCCACAGGCCCCUCGGGGAUGU----AUCUCCCCGAGC .((((((......((((....))))....((((...((.....))...)))).......))))))........((((((((...----...)))))))). ( -27.50) >DroSec_CAF1 20634 92 + 1 AGGGAAAUGCCCUGCAGCUAAC----GAAUGUUAAAUGGUAAUCAAGAAACAAUCAAACUUUCCCACAGGCCCCUCGGGGAUGU----AACUCCCCGAGC .((((((((((.(..(((....----....)))..).)))).....((.....))....))))))........((((((((...----...)))))))). ( -26.20) >DroSim_CAF1 20456 92 + 1 AGGGAAAUGCCCUGCAGCUAAC----GAAUGUUAAAUGGUAAUCAAGAAACAAUCAAACUUUCCCACAGGCCCCUCGGGGAUGU----AACUCCCCGAGC .((((((((((.(..(((....----....)))..).)))).....((.....))....))))))........((((((((...----...)))))))). ( -26.20) >DroEre_CAF1 21328 92 + 1 AGGGAAAUGCCCUGCAGCUAAC----GAAUGUUAAAUGGUAAUCAAGAAACAAUCAAACUUUCCCACAGCCCCCUCGGGGAUGU----AUCUCCCCGAAC .((((((((((.(..(((....----....)))..).)))).....((.....))....)))))).........(((((((...----...))))))).. ( -23.80) >DroYak_CAF1 23154 96 + 1 AGGGAAAUGCCCUGCAGCUAAC----GAAUGUUAAAUGGUAAUCAAGAAACAAUCAAACUUUCCCACAGGCCCCUCGGGCAUGUACAUAUCUCCCCGAGC .((((((((((.(..(((....----....)))..).)))).....((.....))....))))))........((((((...(.......)..)))))). ( -19.60) >consensus AGGGAAAUGCCCUGCAGCUAAC____GAAUGUUAAAUGGUAAUCAAGAAACAAUCAAACUUUCCCACAGGCCCCUCGGGGAUGU____AUCUCCCCGAGC .((((((((((.(..(((............)))..).)))).....((.....))....))))))........((((((((.(.......))))))))). (-22.06 = -22.46 + 0.40)

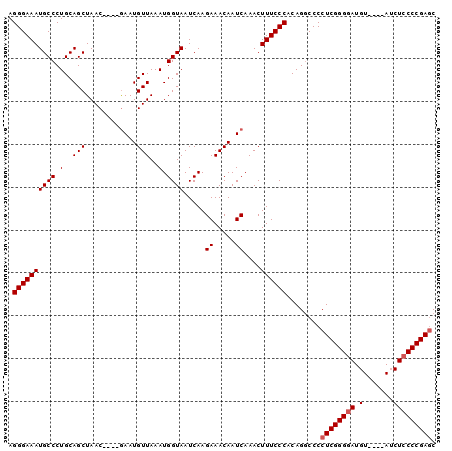
| Location | 4,256,247 – 4,256,343 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.44 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4256247 96 - 23771897 GCUCGGGGAGAU----ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACAUUUAGCAGUUAGCUGCAGGGCAUUUCCCU .((((((((...----...))))))))........((((((((((((.((((...((.....))...)))).)))((((....))))..))).)))))). ( -34.00) >DroSec_CAF1 20634 92 - 1 GCUCGGGGAGUU----ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACAUUC----GUUAGCUGCAGGGCAUUUCCCU .((((((((...----...)))))))).(.((((..(.......(((.((((...((.....))...)))).))----)....)..)))).)........ ( -31.80) >DroSim_CAF1 20456 92 - 1 GCUCGGGGAGUU----ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACAUUC----GUUAGCUGCAGGGCAUUUCCCU .((((((((...----...)))))))).(.((((..(.......(((.((((...((.....))...)))).))----)....)..)))).)........ ( -31.80) >DroEre_CAF1 21328 92 - 1 GUUCGGGGAGAU----ACAUCCCCGAGGGGGCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACAUUC----GUUAGCUGCAGGGCAUUUCCCU ..(((((((...----...)))))))(((((((.((....(((((((.((((...((.....))...)))).))----)..)))).)).)))...)))). ( -29.40) >DroYak_CAF1 23154 96 - 1 GCUCGGGGAGAUAUGUACAUGCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACAUUC----GUUAGCUGCAGGGCAUUUCCCU .((((((.(..........).)))))).(.((((..(.......(((.((((...((.....))...)))).))----)....)..)))).)........ ( -26.00) >consensus GCUCGGGGAGAU____ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACAUUC____GUUAGCUGCAGGGCAUUUCCCU .((((((((..........))))))))(((((((..(.((.....((.((((...((.....))...)))).)).....))..)..))))......))). (-27.36 = -27.44 + 0.08)



| Location | 4,256,276 – 4,256,383 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -32.60 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4256276 107 - 23771897 CCCAUGGGCUAUUAUCUCGACCCCGGCUGUAGGUGCAUUUGCUCGGGGAGAU----ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACA ((((((((((..(((((((.(....).)).)))))......((((((((...----...)))))))).))))))))))...(((.(((.((.........)).))).))). ( -37.80) >DroSec_CAF1 20659 107 - 1 CCCAUGGGCCAUUAUCUCGACCUCGGCUGUAGGUGCAUUUGCUCGGGGAGUU----ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACA ((((((((((..(((((((.(....).)).)))))......((((((((...----...)))))))).))))))))))...(((.(((.((.........)).))).))). ( -40.10) >DroSim_CAF1 20481 107 - 1 CCCAUGGGCCAUUAUCUCGACCUCGGCUGUAGGUGCAUUUGCUCGGGGAGUU----ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACA ((((((((((..(((((((.(....).)).)))))......((((((((...----...)))))))).))))))))))...(((.(((.((.........)).))).))). ( -40.10) >DroEre_CAF1 21353 107 - 1 CCCAGGGGCUAUAGUCUCGACCUCGGCUGUAAGUGCAUUUGUUCGGGGAGAU----ACAUCCCCGAGGGGGCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACA (((((((((....)))))..((((.((.......))......(((((((...----...)))))))))))....))))...(((.(((.((.........)).))).))). ( -33.00) >DroYak_CAF1 23179 111 - 1 CCCAGGGGCUAUAGUCUCGACCUCGGCUGUAGGUGCAAUUGCUCGGGGAGAUAUGUACAUGCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACA ((((.(((((..........((((((..(((.(((((....(((...)))...))))).)))))))))))))).))))...(((.(((.((.........)).))).))). ( -34.30) >consensus CCCAUGGGCUAUUAUCUCGACCUCGGCUGUAGGUGCAUUUGCUCGGGGAGAU____ACAUCCCCGAGGGGCCUGUGGGAAAGUUUGAUUGUUUCUUGAUUACCAUUUAACA ((((((((((...............((.......)).....((((((((..........)))))))).))))))))))...(((.(((.((.........)).))).))). (-32.60 = -33.00 + 0.40)

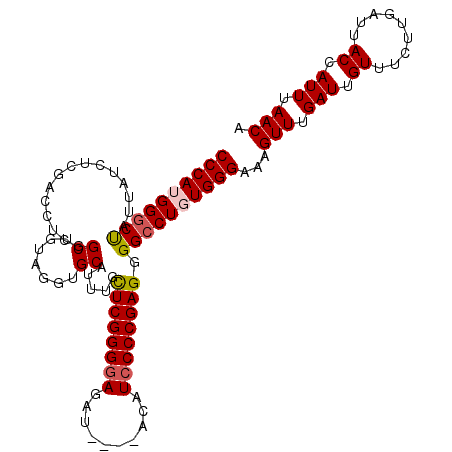

| Location | 4,256,307 – 4,256,409 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -31.70 |
| Energy contribution | -32.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4256307 102 - 23771897 --CUCUUAAUUACAAAUAAUCAAUG------------AACCCCAUGGGCUAUUAUCUCGACCCCGGCUGUAGGUGCAUUUGCUCGGGGAGAU----ACAUCCCCGAGGGGCCUGUGGGAA --.......................------------...((((((((((..(((((((.(....).)).)))))......((((((((...----...)))))))).)))))))))).. ( -36.90) >DroSec_CAF1 20690 116 - 1 UCCUCUUAAUUACAAAUAAUCAAUGACUUAUGUUCAGAACCCCAUGGGCCAUUAUCUCGACCUCGGCUGUAGGUGCAUUUGCUCGGGGAGUU----ACAUCCCCGAGGGGCCUGUGGGAA ........................................((((((((((..(((((((.(....).)).)))))......((((((((...----...)))))))).)))))))))).. ( -39.20) >DroSim_CAF1 20512 116 - 1 UCCUCUUAAUUACAAAUAAUCAAUGACUUAUGUUCAGAGCCCCAUGGGCCAUUAUCUCGACCUCGGCUGUAGGUGCAUUUGCUCGGGGAGUU----ACAUCCCCGAGGGGCCUGUGGGAA ..((((.(((.....................))).)))).((((((((((..(((((((.(....).)).)))))......((((((((...----...)))))))).)))))))))).. ( -40.60) >DroEre_CAF1 21384 114 - 1 --CCUUUAAUUAACAAUAAUCAAUGACUUAUGUUCAGAACCCCAGGGGCUAUAGUCUCGACCUCGGCUGUAAGUGCAUUUGUUCGGGGAGAU----ACAUCCCCGAGGGGGCUGUGGGAA --(((...................(((....)))(((..((((..(.(((((((((........)))))).))).)......(((((((...----...))))))))))).))).))).. ( -34.20) >DroYak_CAF1 23210 118 - 1 --CUUCUAAUUAACGACAAUCAAUGACUUAUGUUUUGAGCCCCAGGGGCUAUAGUCUCGACCUCGGCUGUAGGUGCAAUUGCUCGGGGAGAUAUGUACAUGCCCGAGGGGCCUGUGGGAA --...........(.(((.((((.(((....)))))))(((((.(((.((((((((........))))))))(((((....(((...)))...)))))...)))..))))).))).)... ( -37.60) >consensus __CUCUUAAUUACAAAUAAUCAAUGACUUAUGUUCAGAACCCCAUGGGCUAUUAUCUCGACCUCGGCUGUAGGUGCAUUUGCUCGGGGAGAU____ACAUCCCCGAGGGGCCUGUGGGAA ........................................((((((((((...............((.......)).....((((((((..........)))))))).)))))))))).. (-31.70 = -32.10 + 0.40)


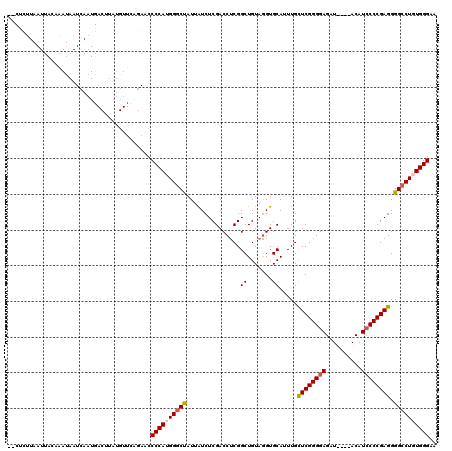
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:27 2006