| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 451,447 – 451,552 |
| Length | 105 |
| Max. P | 0.749008 |

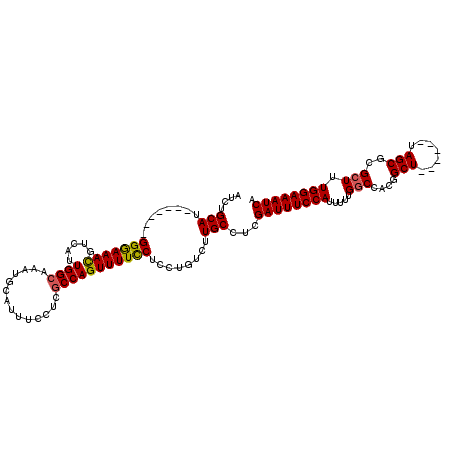
| Location | 451,447 – 451,552 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.11 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 451447 105 + 23771897 AUCUGCAU-------GGGAAACGUCAUUGGCAAAUGCAUUUCCUCGCCAGUUUUCCUCCUGUCUUGCCUCGAUUUCCAUUUUUGGCCACGGCU------UAGCGCGAUUUGGAAAUCA ....(((.-------((((((....((((((..............))))))))))).).)))........((((((((..((.(((....)))------......))..)))))))). ( -24.34) >DroSec_CAF1 32420 111 + 1 AUAUGCAU-------GGGAAAUGUCAUUGGCAAAUGCAUUUCCUCGCCAGUUUUCCUCCUGUCUUGCCUCGAUUUCCAUUUUUGGCCACGGCUUAGGCUUAGCCCGCUUUGGAAAUCA ....(((.-------((((((((.((((....)))))))))))).(.(((........))).).)))...((((((((.....(((...(((....)))..))).....)))))))). ( -32.80) >DroSim_CAF1 32774 105 + 1 AUCUGCAU-------GGGAAACGUCAUUGGCAAAUACAUUUCCUCGCCAGUUUUCCUCCUGUCUUGCCUCGAUUUCCAUUUUUGGCCACGGCU------UAGCGCGCUUUGGAAAUCA ....(((.-------((((((....((((((..............))))))))))).).)))........((((((((.....((((.(....------..).).))).)))))))). ( -24.44) >DroEre_CAF1 34496 104 + 1 AUCUGCAU-------GGAAAACGUCAUUGGAAAAGGCAUUUCCUCGCCAGUUUUUCCCCG-UCUUGCCACGAUUUCCAUUUUUGGCCACGGCU------UAGCGCGCUUUGGAAAUCA .......(-------((((.(((.....(((((((((........)))...)))))).))-).)).))).((((((((.....((((.(....------..).).))).)))))))). ( -24.70) >DroYak_CAF1 33697 112 + 1 AUCUGCAUGGAGCAUGGAAAACGUCAUUGGCAAAUGCAUUUCCUCGCCAGUUUUCCUCCACUCUUGCCUCGAUUUCCAUUUUUGGCCGCGGCU------UAGCGCGCUUUGGAAAUCA ....(((.((((...(((((((.....((((..............)))))))))))....)))))))...((((((((.....((((((....------..))).))).)))))))). ( -35.94) >consensus AUCUGCAU_______GGGAAACGUCAUUGGCAAAUGCAUUUCCUCGCCAGUUUUCCUCCUGUCUUGCCUCGAUUUCCAUUUUUGGCCACGGCU______UAGCGCGCUUUGGAAAUCA ....(((........(((((((.....((((..............)))))))))))........)))...((((((((.....(((....(((.......)))..))).)))))))). (-23.27 = -23.11 + -0.16)

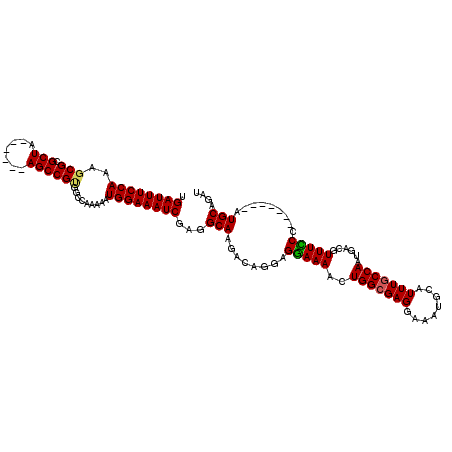
| Location | 451,447 – 451,552 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -23.93 |
| Energy contribution | -23.81 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 451447 105 - 23771897 UGAUUUCCAAAUCGCGCUA------AGCCGUGGCCAAAAAUGGAAAUCGAGGCAAGACAGGAGGAAAACUGGCGAGGAAAUGCAUUUGCCAAUGACGUUUCCC-------AUGCAGAU .((((((((......((((------.....))))......))))))))...(((...(((........)))..(.(((((((((((....)))).))))))))-------.))).... ( -28.60) >DroSec_CAF1 32420 111 - 1 UGAUUUCCAAAGCGGGCUAAGCCUAAGCCGUGGCCAAAAAUGGAAAUCGAGGCAAGACAGGAGGAAAACUGGCGAGGAAAUGCAUUUGCCAAUGACAUUUCCC-------AUGCAUAU .((((((((.....(((((.((....))..))))).....))))))))...(((...(((........)))..(.(((((((((((....)))).))))))))-------.))).... ( -35.30) >DroSim_CAF1 32774 105 - 1 UGAUUUCCAAAGCGCGCUA------AGCCGUGGCCAAAAAUGGAAAUCGAGGCAAGACAGGAGGAAAACUGGCGAGGAAAUGUAUUUGCCAAUGACGUUUCCC-------AUGCAGAU .((((((((..((.(((..------....)))))......))))))))...(((...(((........)))..(.((((((((...........)))))))))-------.))).... ( -26.60) >DroEre_CAF1 34496 104 - 1 UGAUUUCCAAAGCGCGCUA------AGCCGUGGCCAAAAAUGGAAAUCGUGGCAAGA-CGGGGAAAAACUGGCGAGGAAAUGCCUUUUCCAAUGACGUUUUCC-------AUGCAGAU .((((((((..((.(((..------....)))))......))))))))((((.((((-((.((((((...((((......)))))))))).....))))))))-------))...... ( -31.30) >DroYak_CAF1 33697 112 - 1 UGAUUUCCAAAGCGCGCUA------AGCCGCGGCCAAAAAUGGAAAUCGAGGCAAGAGUGGAGGAAAACUGGCGAGGAAAUGCAUUUGCCAAUGACGUUUUCCAUGCUCCAUGCAGAU .((((((((..((.(((..------....)))))......))))))))...(((.(((((..((((((((((((((........))))))).....))))))).)))))..))).... ( -37.20) >consensus UGAUUUCCAAAGCGCGCUA______AGCCGUGGCCAAAAAUGGAAAUCGAGGCAAGACAGGAGGAAAACUGGCGAGGAAAUGCAUUUGCCAAUGACGUUUCCC_______AUGCAGAU .((((((((..(((.(((.......)))))).........))))))))...(((........(((((..(((((((........)))))))......))))).........))).... (-23.93 = -23.81 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:47 2006