
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,201,062 – 4,201,165 |
| Length | 103 |
| Max. P | 0.853527 |

| Location | 4,201,062 – 4,201,165 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4201062 103 + 23771897 CAGUGACGUGCUGGAUCUUACCGAUCCAAAAUCGUUCCGUGACCUAUCCAAACCCAUUGGAUGCGUAAGUAUUA-CU--GAAAAUAGAAAAAGGUGAUCAG-UUACA ..(((((....((((((.....))))))..........((.((((((((((.....))))))((....))....-..--............)))).))..)-)))). ( -25.00) >DroPse_CAF1 48755 102 + 1 UAGUGAUGUGCUGGAUCUGACGGAUCCGAAAUCAUUCCGAGACUUGUCCAAGCCCAUUGGAUGUGUAAGUGUUGAAU--GAUUCAUAAAUGA---AAUCGCAUUAAA ...(((((((..(((((.....)))))(((.((((((.((.((((((((((.....))))))....)))).))))))--)))))........---...))))))).. ( -29.90) >DroGri_CAF1 52640 102 + 1 CAGUGACGUGCUGGAUCUAACCGAUCCUCGUUCAUUUCGUGACUUGUCCAAGCCAAUUGGAUGCGUAAGUCUCUUCUAAAAUAAUAAAGUAU---CAUUUG-UUA-A .(((((((....(((((.....))))).)).)))))..(.(((((((((((.....))))))....))))).)........(((((((....---..))))-)))-. ( -23.40) >DroEre_CAF1 48736 103 + 1 CAGUGACGUGCUGGAUCUAACCGAUCCAAAAUCAUUCCGUGACCUUUCCAAACCCAUUGGAUGCGUAAGUAUUA-CU--GACCUUGAAAUGAGAUGAUCAC-UUACA ..((((.(((.((((((.....))))))..(((((..(((......(((((.....)))))(((....)))...-..--.........)))..))))))))-)))). ( -24.90) >DroYak_CAF1 53438 103 + 1 CAGUGACGUGCUGGAUCUAACCGAUCCCAAAUCAUUCCGUGACCUAUCCAAACCAAUUGGAUGCGUAAGUAUCG-CU--GACCUCAAAAUAAGGUGAUUAG-UUACA .(((((..((..(((((.....)))))))..)))))..(((((..(((...........(((((....))))).-..--.((((.......)))))))..)-)))). ( -26.50) >DroPer_CAF1 49441 102 + 1 UAGUGAUGUGCUGGAUCUGACGGAUCCGAAAUCAUUCCGAGACUUGUCCAAGCCCAUUGGAUGUGUAAGUGUUGAAU--GAUUCAUAAAUGA---AAACGCAUUAAA ...(((((((..(((((.....)))))(((.((((((.((.((((((((((.....))))))....)))).))))))--)))))........---...))))))).. ( -30.30) >consensus CAGUGACGUGCUGGAUCUAACCGAUCCAAAAUCAUUCCGUGACCUGUCCAAACCCAUUGGAUGCGUAAGUAUUG_CU__GAUAAUAAAAUAA___GAUCAC_UUACA ........(((((((((.....)))))..............((.(((((((.....))))))).)).)))).................................... (-14.42 = -14.42 + -0.00)

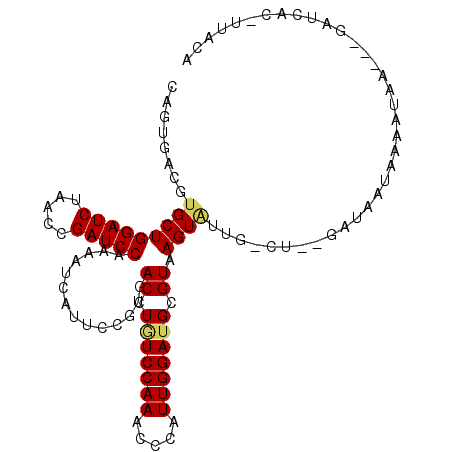
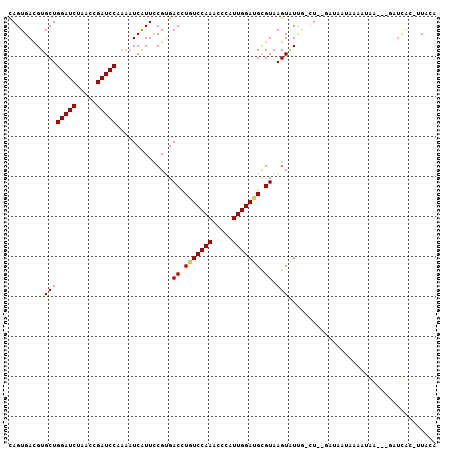
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:07 2006