
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,197,139 – 4,197,246 |
| Length | 107 |
| Max. P | 0.999671 |

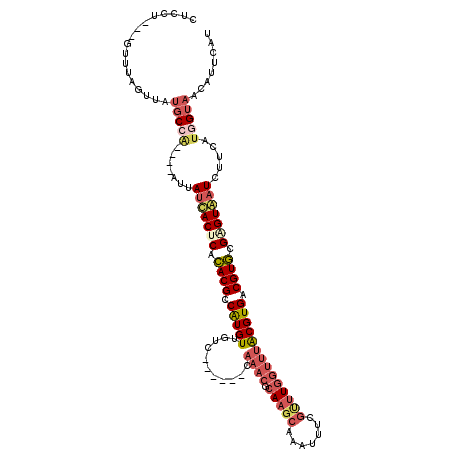
| Location | 4,197,139 – 4,197,246 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.19 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -13.46 |
| Energy contribution | -16.40 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4197139 107 + 23771897 CUGCU---GUUAAGUUAUGCCG----AUUAUCACUCACACGCCAUGUUGUC------CAAACGCAAGCAAAUUUCGCUUGGUUUGCGUGACGUGCGAGUAAUCUUCAUAGUAACAUACAU ....(---((((...((((..(----(((...((((.((((.((((.....------(((((.(((((.......)))))))))))))).)))).))))))))..)))).)))))..... ( -31.20) >DroSec_CAF1 46976 107 + 1 CUCCU---GUUUAGUUAUGCCA----AUUAUUACUCACACGCCAUGUUGUC------CAAACGCAAGCAAAUUUCGUUUGGUUUACGUGACGUGCGGGUAAUCUUCAUGGUAACAUUCAU .....---.........(((((----...(((((((.((((.(((((...(------((((((.((.....)).)))))))...))))).)))).))))))).....)))))........ ( -33.50) >DroSim_CAF1 47193 107 + 1 CUCCU---GUUUAGUUAUGCCA----AUUAUUACUCACACGCCAUGUUGUC------CAAACGCAAGCAAAUUUCGUUUGGUUUACGUGACGUGCGUGUAAUCUUCAUGGUAACAUUCAU .....---.........(((((----...(((((.(.((((.(((((...(------((((((.((.....)).)))))))...))))).)))).).))))).....)))))........ ( -30.20) >DroAna_CAF1 46013 108 + 1 CUUUUAUAUUUUCAUCAAGCCAUUAGAAUAUCACGCAUACGCCGUGUGUUCUUGGCUUGAAGGCAUGAAAAC------UAGCCAACGUGUCGUAUGAGUGUUUUUU--UGAAGCAA---- ..(((((((.....((((((((..((((((.(((((....).)))))))))))))))))).((((((.....------.......)))))))))))))((((((..--.)))))).---- ( -30.70) >consensus CUCCU___GUUUAGUUAUGCCA____AUUAUCACUCACACGCCAUGUUGUC______CAAACGCAAGCAAAUUUCGUUUGGUUUACGUGACGUGCGAGUAAUCUUCAUGGUAACAUUCAU .................(((((.......(((((((.((((.(((((...........((((.(((((.......)))))))))))))).)))).))))))).....)))))........ (-13.46 = -16.40 + 2.94)


| Location | 4,197,139 – 4,197,246 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.19 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4197139 107 - 23771897 AUGUAUGUUACUAUGAAGAUUACUCGCACGUCACGCAAACCAAGCGAAAUUUGCUUGCGUUUG------GACAACAUGGCGUGUGAGUGAUAAU----CGGCAUAACUUAAC---AGCAG .(((..((((...(((..(((((((((((((((.(((((((((((((...))))))).)))))------.....).)))))))))))))))..)----))...))))...))---).... ( -39.10) >DroSec_CAF1 46976 107 - 1 AUGAAUGUUACCAUGAAGAUUACCCGCACGUCACGUAAACCAAACGAAAUUUGCUUGCGUUUG------GACAACAUGGCGUGUGAGUAAUAAU----UGGCAUAACUAAAC---AGGAG ......(((((((.....(((((.(((((((((.((...(((((((...........))))))------)...)).))))))))).)))))...----)))..)))).....---..... ( -31.60) >DroSim_CAF1 47193 107 - 1 AUGAAUGUUACCAUGAAGAUUACACGCACGUCACGUAAACCAAACGAAAUUUGCUUGCGUUUG------GACAACAUGGCGUGUGAGUAAUAAU----UGGCAUAACUAAAC---AGGAG ......(((((((.....(((((.(((((((((.((...(((((((...........))))))------)...)).))))))))).)))))...----)))..)))).....---..... ( -31.50) >DroAna_CAF1 46013 108 - 1 ----UUGCUUCA--AAAAAACACUCAUACGACACGUUGGCUA------GUUUUCAUGCCUUCAAGCCAAGAACACACGGCGUAUGCGUGAUAUUCUAAUGGCUUGAUGAAAAUAUAAAAG ----....((((--.......................(((..------........))).((((((((((((..((((.......))))...))))..)))))))))))).......... ( -22.60) >consensus AUGAAUGUUACCAUGAAGAUUACUCGCACGUCACGUAAACCAAACGAAAUUUGCUUGCGUUUG______GACAACAUGGCGUGUGAGUAAUAAU____UGGCAUAACUAAAC___AGGAG ....((((((........(((((.((((((((..(((((..........)))))....(((........))).....)))))))).))))).......))))))................ (-16.72 = -17.29 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:05 2006