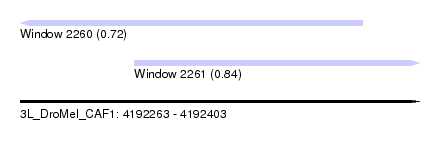
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,192,263 – 4,192,403 |
| Length | 140 |
| Max. P | 0.836112 |

| Location | 4,192,263 – 4,192,383 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
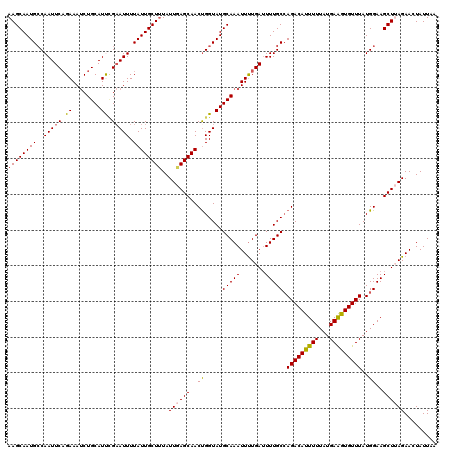
>3L_DroMel_CAF1 4192263 120 - 23771897 AAGCAAUGCCAAUUCAGAAAUCUGCAUUCGAAUUUUAUUGCUUUAUUGAGCAACUGGUAUGCAAAUUUUGAUUUUGCCAGACAUUUUUAUGAAGUGUUUAUGGAAGCUUAGAACUAUUAA ((((((((..(((((.(((.......)))))))).))))))))..((((((..((((((..((.....))....))))))(((((((...)))))))........))))))......... ( -25.10) >DroSec_CAF1 42164 120 - 1 AAGCAAUGCCAAUUCAGAAAUCUGCAUUCGAAUUUUAUUGCUUUAUUAAGCAACUGGUAUGCAAAUUUUGAUUUUGCCAGACAUUUUUAUGAAGUGUUUAUGGAAGCUAAGAACUAUUAA .(((.((((((...(((....))).............((((((....)))))).))))))(((((.......))))).(((((((((...)))))))))......)))............ ( -23.30) >DroSim_CAF1 42360 120 - 1 AAGCAAUGCCAAUUCAGAAAUCUGCAUUCGAAUUUUAUUGCUUUAUUAAGCAACUGGUAUGCAAAUUUUGAUUUUGCCAGACAUUUUUAUGAAGUGUUUAUGGAAGCUAAGAACUAUUAA .(((.((((((...(((....))).............((((((....)))))).))))))(((((.......))))).(((((((((...)))))))))......)))............ ( -23.30) >DroEre_CAF1 38572 119 - 1 AAGCAAUGGCAAUUCCGAAAUCUGCAUCUGAAUUUUAUUGCU-UAUUGAGCAACCGGUAUGCAAAUUUUGAUUUUGCGAGACAUUUUUAUGAAGUGUUUAUGGAAGCUUAGAACUAUUAA (((((((((.(((((.((........)).)))))))))))))-).((((((..(((...((((((.......))))))(((((((((...))))))))).)))..))))))......... ( -30.20) >DroYak_CAF1 44165 120 - 1 AAGCAAUGCCAAUUCAGAAAACUGCAUCAGCAUUUUAUUGCUUUAUUGAGCAACUGGUAUGCAAAUUCUGAGUUUGCCAGACACUUUUAUGAGGUGUUUAUGGAAGCUUAGAACUAUUAA ..(((.(((((.......(((.(((....))).))).((((((....)))))).))))))))...((((((((((.(((((((((((...))))))))..)))))))))))))....... ( -36.60) >consensus AAGCAAUGCCAAUUCAGAAAUCUGCAUUCGAAUUUUAUUGCUUUAUUGAGCAACUGGUAUGCAAAUUUUGAUUUUGCCAGACAUUUUUAUGAAGUGUUUAUGGAAGCUUAGAACUAUUAA ((((((((..(((((.((........)).))))).))))))))..((((((..((.....(((((.......))))).(((((((((...)))))))))..))..))))))......... (-21.36 = -21.44 + 0.08)


| Location | 4,192,303 – 4,192,403 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.36 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
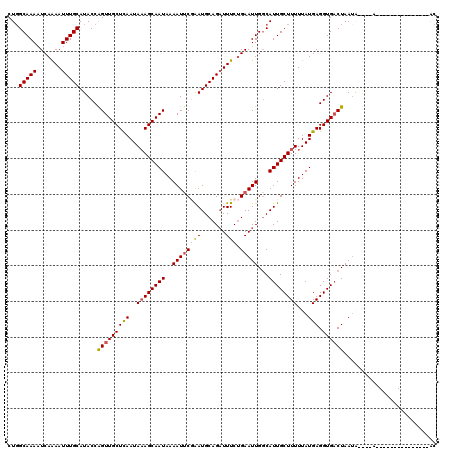
>3L_DroMel_CAF1 4192303 100 + 23771897 CUGGCAAAAUCAAAAUUUGCAUACCAGUUGCUCAAUAAAGCAAUAAAAUUCGAAUGCAGAUUUCUGAAUUGGCAUUGCUUUUUAUGAGGUGACUAAUA--------------------AC ...(((((.......))))).....(((..((((..((((((((..((((((((.......))).)))))...))))))))...))).)..)))....--------------------.. ( -23.10) >DroSec_CAF1 42204 100 + 1 CUGGCAAAAUCAAAAUUUGCAUACCAGUUGCUUAAUAAAGCAAUAAAAUUCGAAUGCAGAUUUCUGAAUUGGCAUUGCUUUUUAUGAGGUGACUAAUA--------------------AC ...(((((.......))))).....(((..(((...((((((((..((((((((.......))).)))))...)))))))).....)))..)))....--------------------.. ( -23.10) >DroSim_CAF1 42400 102 + 1 CUGGCAAAAUCAAAAUUUGCAUACCAGUUGCUUAAUAAAGCAAUAAAAUUCGAAUGCAGAUUUCUGAAUUGGCAUUGCUUUUUAUGAGGUGACUAAUA----A--------------AAC .((.(((..(((((((((((((....(((((((....))))))).........)))))))))).))).))).)).....((((((.((....)).)))----)--------------)). ( -23.12) >DroEre_CAF1 38612 105 + 1 CUCGCAAAAUCAAAAUUUGCAUACCGGUUGCUCAAUA-AGCAAUAAAAUUCAGAUGCAGAUUUCGGAAUUGCCAUUGCUUUUUAUGAGGUGUCUAAUGGGCUA--------------UAC ...((....((.((((((((((....((((((.....-)))))).........)))))))))).)).....((((((((((....)))))....)))))))..--------------... ( -22.52) >DroYak_CAF1 44205 120 + 1 CUGGCAAACUCAGAAUUUGCAUACCAGUUGCUCAAUAAAGCAAUAAAAUGCUGAUGCAGUUUUCUGAAUUGGCAUUGCUUUUUAUGAGGUGACUAAGUGACUAAGGUGACUAUGCAGUGC ((((.....))))...((((((...(((..((((..((((((.....((((..((.(((....))).))..))))))))))...))).)..))).(((.((....)).)))))))))... ( -33.50) >consensus CUGGCAAAAUCAAAAUUUGCAUACCAGUUGCUCAAUAAAGCAAUAAAAUUCGAAUGCAGAUUUCUGAAUUGGCAUUGCUUUUUAUGAGGUGACUAAUA____A_______________AC ...(((((.......))))).....(((((((((..((((((((..((((((((.......))).)))))...))))))))...))).)))))).......................... (-18.00 = -18.36 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:03 2006