| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 448,377 – 448,479 |
| Length | 102 |
| Max. P | 0.747899 |

| Location | 448,377 – 448,479 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -13.94 |
| Energy contribution | -13.58 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
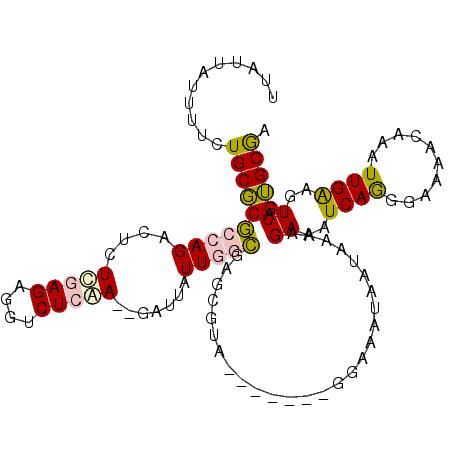
>3L_DroMel_CAF1 448377 102 + 23771897 UUAUUAUUUUCUGCGCGCCAGACUCUUGAGAGGUCUCAA--GAUUAUUGGCGAGCGUA-------GGAAAUAAUAAAAGAAAUCAGGGAAAACAAAUUGAAGUCAGUGCGA (((((((((((((((((((((..(((((((....)))))--))...)))))..)))))-------)))))))))))..((..((((..........))))..))....... ( -36.60) >DroPse_CAF1 86000 102 + 1 ---------CCUGCUCUAGAGACAUUCAAGAGGUCUCGAGCGAUUAUUGGGGAGCGUACGGUACGGGAAAUAAUAAAAGAAAUCAAGGCAAACAAAUUGAAGUCAGGGCAA ---------..((((((...(((.(((((...((((.((.(..((((((.....((((...)))).....))))))..)...)).)))).......)))))))))))))). ( -22.10) >DroSim_CAF1 29726 102 + 1 UUAUUAUUUUCUGCGCGCCAGACUCUUGAGAGGUCUCAA--GAUUAUUGGCGAGCGUA-------GGAAAUAAUAAAAGAAAUCAGGGAAAACAAAUUGGAGUCAGUGCGA (((((((((((((((((((((..(((((((....)))))--))...)))))..)))))-------)))))))))))..((..((((..........))))..))....... ( -35.70) >DroEre_CAF1 31405 102 + 1 UUAUUAUUUUCUGCGCGCCAGCCUCUCGAGAGGUCUCAA--GAUUAUUGGCGAGCGUA-------GGAAAUAAUAAAAGAAAUCAGGGAAAACAAAUUGAAGUCAGUGCGA (((((((((((((((((((((..(((.(((....))).)--))...)))))..)))))-------)))))))))))..((..((((..........))))..))....... ( -32.50) >DroYak_CAF1 30492 102 + 1 GUAUUAUUUUCUGCGCGCCAGACUCUCGAGAGGUCUCAA--GAUUAUUGGCGGGCGUA-------GGAAAUAAUAAGAGAAAUCAGGGAAAACAAAUUGAAGUCAGUGCGA .((((((((((((((((((((..(((.(((....))).)--))...)))))..)))))-------)))))))))).((....))...........((((....)))).... ( -32.40) >DroPer_CAF1 83924 102 + 1 ---------CCUGCUCUAGAGACAUUCAAGAGGUCUCGAGCGAUUAUUGGGGAGCGUACGGUACGGGAAAUAAUAAAAGAAAUCAAGGCAAACAAAUUGAAGUCAGGGCAA ---------..((((((...(((.(((((...((((.((.(..((((((.....((((...)))).....))))))..)...)).)))).......)))))))))))))). ( -22.10) >consensus UUAUUAUUUUCUGCGCGCCAGACUCUCGAGAGGUCUCAA__GAUUAUUGGCGAGCGUA_______GGAAAUAAUAAAAGAAAUCAGGGAAAACAAAUUGAAGUCAGUGCGA ...........((((((((((....(((((....))))).......)))))...........................((..((((..........))))..)).))))). (-13.94 = -13.58 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:44 2006