
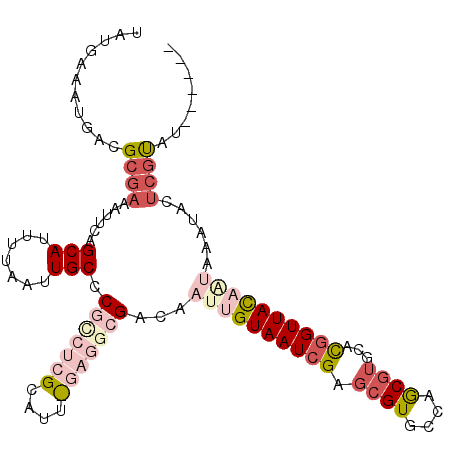
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,189,298 – 4,189,428 |
| Length | 130 |
| Max. P | 0.883773 |

| Location | 4,189,298 – 4,189,402 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -16.62 |
| Energy contribution | -19.18 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4189298 104 + 23771897 UAUGAAAUGACGCGAAAUUCAGCAUUUUAAUUGCCCGCCUCGCAUUUGAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCAUGGUUAUAAUAAAUACUCGCAU------ ...........((((......(((.......))).(((((((....)))))))...((((((((((.(((((....))))).))))))))))......))))..------ ( -30.30) >DroPse_CAF1 36172 94 + 1 UAUGAAAUGACGCGAAAUCCAGCAUUUUAAUUGCUC--CUC---------GCGACAAUUGUAAUCGAGCGUGCCAACGUCUAUGGUUAC-----AUACUCGUCUAAGUAC ........(((((((.....((((.......)))).--.))---------))((....(((((((((((((....))).)).)))))))-----)...)))))....... ( -22.50) >DroSec_CAF1 34902 104 + 1 UAUGAAAUGACGAAAAAUUCAGCAUUUUAAUUGCCCGUCUCGCAUUCGAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCACGGUUAUAAUAAAUACUCGUAU------ ..(((((((..((.....))..)))))))..(((.(((((((....)))))))...((((((((((.(((((....))))).))))))))))........))).------ ( -28.10) >DroEre_CAF1 35474 100 + 1 UAUGAAAUGACGCGAAAUUCAGCAUUUUAAUUGCCCGCCUCGCAUUUGAGGCGACAAUUGUAAUCGAGCGUGCCAGC----UCCGUUACAGUAAAUACACGUAU------ .........(((.(.......(((.......))).(((((((....)))))))...((((((((.((((......))----)).)))))))).....).)))..------ ( -28.90) >DroYak_CAF1 39584 104 + 1 UAUGAAAUGACGCGAAAUUCAGCAUUUUAAUUGCCCGCCUCGCAUUCGAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCACGGUUACAAUAAAUACUCGCAU------ ...........((((......(((.......))).(((((((....)))))))...((((((((((.(((((....))))).))))))))))......))))..------ ( -35.20) >DroPer_CAF1 36751 94 + 1 UAUGAAAUGACGCGAAAUCCAGCAUUUUAAUUGCUC--CUC---------GCGACAAUUGUAAUCGAGCGUGCCAACGUCUAUGGUUAC-----AUACUCGUCUAAGUAC ........(((((((.....((((.......)))).--.))---------))((....(((((((((((((....))).)).)))))))-----)...)))))....... ( -22.50) >consensus UAUGAAAUGACGCGAAAUUCAGCAUUUUAAUUGCCCGCCUCGCAUU_GAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCACGGUUACAAUAAAUACUCGUAU______ ...........((((......(((.......))).(((((((....)))))))...((((((((((.((((....))))...))))))))))......))))........ (-16.62 = -19.18 + 2.56)


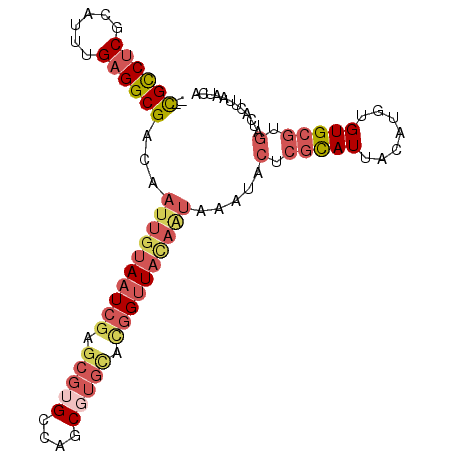
| Location | 4,189,333 – 4,189,428 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -19.16 |
| Energy contribution | -21.48 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4189333 95 + 23771897 --CGCCUCGCAUUUGAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCAUGGUUAUAAUAAAUACUCGCAUUACAUGUGUGCGUGAUCACUUAAUCA --(((((((....)))))))...((((((((((.(((((....))))).))))))))))...(((.(((((.....))))).)))............ ( -29.10) >DroSec_CAF1 34937 95 + 1 --CGUCUCGCAUUCGAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCACGGUUAUAAUAAAUACUCGUAUUACAUGUGUGCGUGAUCACUUAAUCA --(((((((....)))))))...((((((((((.(((((....))))).)))))))))).....(.(((((.......))))).)............ ( -27.10) >DroEre_CAF1 35509 91 + 1 --CGCCUCGCAUUUGAGGCGACAAUUGUAAUCGAGCGUGCCAGC----UCCGUUACAGUAAAUACACGUAUUACAUGUGUGCGUGAUCACUUAAUCA --(((((((....)))))))...((((((((.((((......))----)).)))))))).....(((((((.......)))))))............ ( -32.80) >DroYak_CAF1 39619 95 + 1 --CGCCUCGCAUUCGAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCACGGUUACAAUAAAUACUCGCAUUACAUGUGUGCGUGAUCACUUAAUCA --(((((((....)))))))...((((((((((.(((((....))))).))))))))))...(((.(((((.....))))).)))............ ( -34.00) >DroAna_CAF1 36721 82 + 1 GCUGCCGCUCUUUUGAGGCGACAAUUGUAAUCGAGCGUGCCG-CCUGUGGGGAU--------------CAUUACAGGUGUGCGUGAUCACUUAAUCA (.((((.(......).)))).)....((.(((..(((..(.(-(((((((....--------------..)))))))))..)))))).))....... ( -23.90) >consensus __CGCCUCGCAUUUGAGGCGACAAUUGUAAUCGAGCGUGCCAGCGUGCACGGUUACAAUAAAUACUCGCAUUACAUGUGUGCGUGAUCACUUAAUCA ..((((((......))))))...((((((((((.(((((....))))).)))))))))).....(.(((((.......))))).)............ (-19.16 = -21.48 + 2.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:01 2006