
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,184,528 – 4,184,662 |
| Length | 134 |
| Max. P | 0.717766 |

| Location | 4,184,528 – 4,184,640 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4184528 112 - 23771897 AAGGGCGUCUUCGACACGACGCUGUUCAAGAGUCCCCUCAGCGAUAGGGAGAUGGAGAGAAAUC------UCCAUGGGCUGCGUCAGAACUUGCCAAAAAAUCAUAAACUAUUCUGAA-- ...((((..(((.....(((((.(((((...(((((((.......)))).)))(((((....))------))).))))).))))).)))..)))).......................-- ( -37.40) >DroVir_CAF1 33639 114 - 1 AAAGGCGUCUUUGACACGACGCUCUUCAAGAGUCCCCUCAGCGAUCGGGAAAUGGAUCGUAAGC------UCCAUUUGCUGCGUCAGAACUUGCCAAAAAAUCAUAAACUAUAUUAAAAU ...((((..((((((.(((((((......(((....))))))).)))(.(((((((.(....).------))))))).)...))))))...))))......................... ( -31.30) >DroGri_CAF1 35427 112 - 1 AAAGGCAUCUUCGAUACGACGCUCUUCAAAAGUCCCCAGAGCGAUCGGGAAAUGGAGCGCCAAC------UCCACGCCCUGCGUCAGAAUUUGCCAAAAAAUCACAAACUAUAUUAAA-- ...((((..(((((..(((((((((............)))))).))(((...(((((......)------))))..))).)..)).)))..)))).......................-- ( -31.60) >DroMoj_CAF1 33750 114 - 1 AAAGGCAUCUUCGAUACGACGCUCUUCAAGACUCCCCAGAGCGAUCGCGAAAUGGAGAGAAAGC------UCCUCGCGCUACGUCAGAACUUGCCAAAAAAUCACAAACUAUAUUAACAG ...((((..(((...(((.((((((............))))))..(((((...((((......)------))))))))...)))..)))..))))......................... ( -30.80) >DroAna_CAF1 31288 112 - 1 AAGGGUGUGUUCGACACCACCUUGUUCAAGAGCCCGCUCAGCGACAGGGAGAUGGAGCGGAAGC------UCCAUGGACUGCGUCAGAACUUGCCAAAAAACCACAAACUGUACUGAC-- ...((((.(((((((....(((((((...(((....)))...)))))))..(((((((....))------))))).......))).)))).)))).......................-- ( -38.60) >DroPer_CAF1 31711 118 - 1 AAAGGCGUCUUUGACACGACGCUCUUCAAGAGCCCGCACAGCGACAGGGAGAUGGAAAGGAACCGGAACAUCCAUGUGCUCCGCCAGAACUUGCCAAAAAACCACAAACUAUACUAAA-- ...((((..((((.......((((.....))))..((..(((.(((.(((..(((.......))).....))).))))))..))))))...)))).......................-- ( -24.70) >consensus AAAGGCGUCUUCGACACGACGCUCUUCAAGAGUCCCCACAGCGAUAGGGAAAUGGAGAGAAAGC______UCCAUGCGCUGCGUCAGAACUUGCCAAAAAAUCACAAACUAUACUAAA__ ...((((..((((((....(((((..............)))))........(((((..............))))).......))).)))..))))......................... (-15.38 = -15.38 + 0.00)

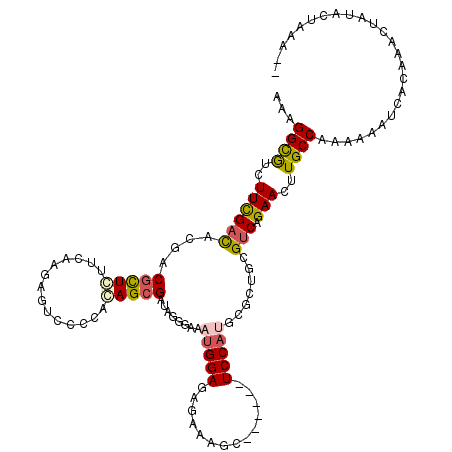

| Location | 4,184,566 – 4,184,662 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.77 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4184566 96 + 23771897 AGCCCAUGGAGAUUUCUCUCCAUCUCCCUAUCGCUGAGGGGACUCUUGAACAGCGUCGUGUCGAAGACGCCCUUAGUAGUCCGCUUGGAUGUGGA-U-- .....(((((((....)))))))....((((..(..((.(((((.((((...(((((........)))))..)))).))))).))..)..)))).-.-- ( -35.00) >DroVir_CAF1 33679 99 + 1 AGCAAAUGGAGCUUACGAUCCAUUUCCCGAUCGCUGAGGGGACUCUUGAAGAGCGUCGUGUCAAAGACGCCUUUCGUGGUGCGCUUCGGCGUCGGGUUU ...(((((((........)))))))(((((.(((((((((.(((..(((((.(((((........))))).))))).))).).)))))))))))))... ( -44.20) >DroGri_CAF1 35465 99 + 1 AGGGCGUGGAGUUGGCGCUCCAUUUCCCGAUCGCUCUGGGGACUUUUGAAGAGCGUCGUAUCGAAGAUGCCUUUGGUGGUGCGCUUCAGUGUCGGAUUU .(((.(((((((....))))))).)))(((.((((..(((.(((.(..(((.(((((........))))))))..).))).).))..)))))))..... ( -37.10) >DroSec_CAF1 30254 96 + 1 AGCCCAUGGAGAUUUCUCUCCAUCUCCCGAUCGCUGAGGGGACUCUUGAACAGCGUCGUGUCGAAGACGCCCUUAGUGGUCCGCUUAGAUGUGGG-U-- .(((((((((((....)))))((((.(.(((((((((((.(((.((.....)).)))(((((...)))))))))))))))).)...)))))))))-)-- ( -39.90) >DroEre_CAF1 30699 96 + 1 ACCCCAUGGAGAUUUCUCUCCAAUUCUCGAUCACUGAGGGGACUCUUGAACAGCGUCGUGUCGAAGACGCCCUUAGUGGUUCGCUUGGAUGUGGG-U-- ..((((((((((....))))).((((.((((((((((((.(((.((.....)).)))(((((...)))))))))))))).)))...)))))))))-.-- ( -37.60) >DroYak_CAF1 34738 96 + 1 AGCCCAUGGAGAUUUCUCUCCAUUUCUCGAUCACUGAGGGGACUCUUGAACAGCGUCGUAUCGAAGACGCCCUUGGUGGUUCGCUUGGAUGUGGG-U-- .(((((((((((....)))))...((((((((((..((((..(....)..).(((((........))))))))..)))).)))...))).)))))-)-- ( -36.60) >consensus AGCCCAUGGAGAUUUCUCUCCAUUUCCCGAUCGCUGAGGGGACUCUUGAACAGCGUCGUGUCGAAGACGCCCUUAGUGGUCCGCUUGGAUGUGGG_U__ .....(((((((....)))))))..(((...(((((((.(((((.((((...(((((........)))))..)))).))))).))))).)).))).... (-26.40 = -26.77 + 0.37)


| Location | 4,184,566 – 4,184,662 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -20.29 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4184566 96 - 23771897 --A-UCCACAUCCAAGCGGACUACUAAGGGCGUCUUCGACACGACGCUGUUCAAGAGUCCCCUCAGCGAUAGGGAGAUGGAGAGAAAUCUCCAUGGGCU --.-(((..((((.((.(((((...((.((((((........)))))).))....))))).))..).)))..))).(((((((....)))))))..... ( -35.10) >DroVir_CAF1 33679 99 - 1 AAACCCGACGCCGAAGCGCACCACGAAAGGCGUCUUUGACACGACGCUCUUCAAGAGUCCCCUCAGCGAUCGGGAAAUGGAUCGUAAGCUCCAUUUGCU ...((((((((.............(((..(((((........)))))..)))..(((....))).))).)))))(((((((.(....).)))))))... ( -36.50) >DroGri_CAF1 35465 99 - 1 AAAUCCGACACUGAAGCGCACCACCAAAGGCAUCUUCGAUACGACGCUCUUCAAAAGUCCCCAGAGCGAUCGGGAAAUGGAGCGCCAACUCCACGCCCU ...........(((((....((......))...)))))....((((((((............)))))).))(((...(((((......)))))..))). ( -24.30) >DroSec_CAF1 30254 96 - 1 --A-CCCACAUCUAAGCGGACCACUAAGGGCGUCUUCGACACGACGCUGUUCAAGAGUCCCCUCAGCGAUCGGGAGAUGGAGAGAAAUCUCCAUGGGCU --.-((((.((((....(((((...((.((((((........)))))).))...).))))(((........)))))))(((((....))))).)))).. ( -33.50) >DroEre_CAF1 30699 96 - 1 --A-CCCACAUCCAAGCGAACCACUAAGGGCGUCUUCGACACGACGCUGUUCAAGAGUCCCCUCAGUGAUCGAGAAUUGGAGAGAAAUCUCCAUGGGGU --.-((((........(((..((((.((((..((((.((((......)))).))))...)))).)))).))).....((((((....)))))))))).. ( -33.90) >DroYak_CAF1 34738 96 - 1 --A-CCCACAUCCAAGCGAACCACCAAGGGCGUCUUCGAUACGACGCUGUUCAAGAGUCCCCUCAGUGAUCGAGAAAUGGAGAGAAAUCUCCAUGGGCU --.-(((.........(((..(((.((.((((((........)))))).))...(((....))).))).)))....(((((((....)))))))))).. ( -31.10) >consensus __A_CCCACAUCCAAGCGAACCACUAAGGGCGUCUUCGACACGACGCUGUUCAAGAGUCCCCUCAGCGAUCGGGAAAUGGAGAGAAAUCUCCAUGGGCU ....((((..((...((........((.((((((........)))))).))...(((....))).))....))....((((((....)))))))))).. (-20.29 = -20.60 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:58 2006